For a “complete” ProteomeXchange submission all the modifications used in the submitted Skyline documents are required to have a Unimod Id. Without Unimod Ids, the data can still be assigned a ProteomeXchange ID, but the data will be annotated as "
supported by repository but incomplete data and/or metadata" when it is announced to ProteomeXchange.
View
Data Validation for ProteomeXchange for more details.
Skyline supports an extensive set of Unimod modifications, and it is recommended that you use modifications from Skyline's built-in list of modifications rather than define your own custom modifications. If you are unable to find your desired modification in Skyline, please post to the
Skyline or
Panorama support boards.
The sections below cover the following:
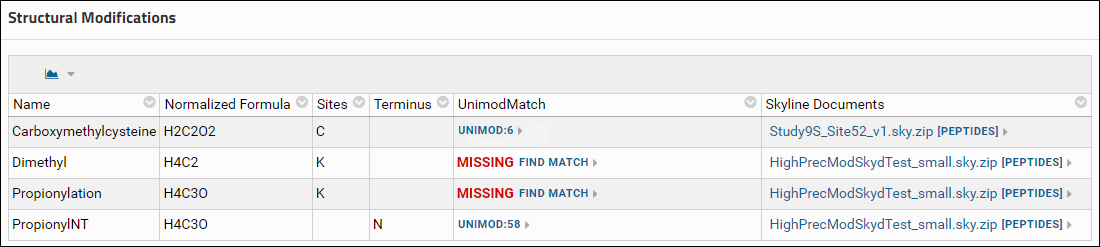
If a modification is not associated with a Unimod Id, you will see a
Find Match link for the modification in two locations:
- If you have already run ProteomeXchange data validation on the data, and are looking at the validation results, you will see the Find Match link in the Unimod Match column of the Modifications table.

- Another location where you will see the Find Match link is on the experiment details page in the Structural Modifications and Isotope Modifications tables. These tables list the structural and isotope modifications used in the dataset. You can go to the experiment details page by clicking the [More Details...] link in the Targeted MS Experiment webpart.
Assign a Unimod match
It is highly recommended that you use a modification from Skyline's built-in list of structural and isotope modifications, since those modifications have a Unimod Id. However, if there are custom modifications in your documents that do not have a Unimod Id, you can find a Unimod match for those modifications on Panorama if the modification definition has a modification formula (or labeled atoms for isotope modifications), the modified sites (amino acids) and / or the modified terminus. To find a Unimod match do the following:
- Click the Find Match link in one of tables (Modifications table on the data validation results page, or one of the modifications table on the experiment details page). If you click this link for a structural modification you will see a page with buttons to find a Unimod Match or define a Combination Modification. Click the button to find a Unimod Match. This will display a page that shows the Unimod matches found for the modification based on the modification formula and modification sites (amino acids) and / or modified terminus. The option to define a combination modification is not available for Isotope modifications, so clicking the Find Match link will open the page with Unimod matches found for the modification.

If more than one match was found, click the
Save Match button below the correct match to associate the Unimod Id with the modification.
Define a combination modification
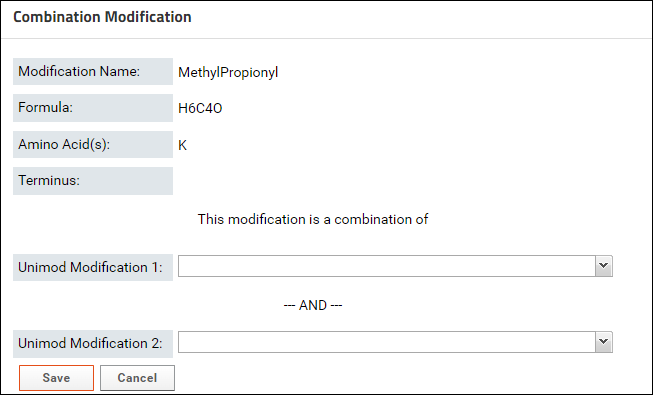
Sometimes users have to create a custom modification in Skyline that is the combination of two modifications. Such modifications do not have a Unimod Id in the Skyline document. To associate a combination modification with the Unimod Ids of the two modifications that it combines, you can do the following:
- Click the Find Match link in one of tables (Modifications table on the data validation results page, or the Structural Modifications table on the experiment details page). You will see the following page:

- Click the button to define a Combination Modification. You will see a form where you can select two Unimod modifications that make up the modification.
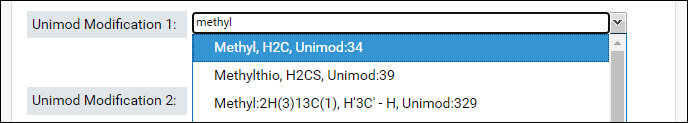
- Start typing the name of first modification in the textbox for Unimod Modification 1. The drop-down list will show you the modifications that match the text entered. Select a modification from the list.
- The sum of the formulas of the two modifications you select must match the formula of combination modification. After you select the first modification, the form will update to show the difference in the formula of the combination modification and the selected modification.

- Select the second modification in Unimod Modification 2. You can search for the modification by name or by the difference in the formula that is displayed in the form (e.g. H4C3O in the image above)
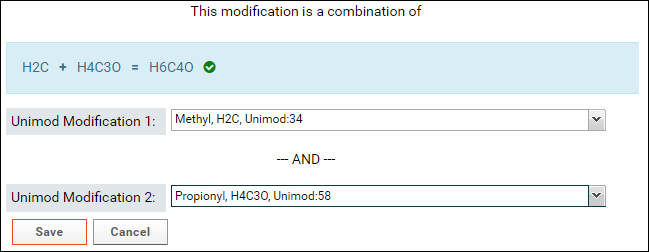
If the sum of the formulas of the two selected modifications matches the formula of the combination modification then a green checkmark will be displayed as shown in the image above.
The screenshot below shows a saved combination modification in the
Modifications table on the data validation results page:

Clicking the Unimod Ids will link to the modification page on the
Unimod website
Wildcard modifications in Skyline
Skyline includes the following built-in wildcard modifications.
- Label:15N
- Label:13C
- Label:13C15N
These modifications implicitly apply to all amino acids. The Unimod modifications corresponding these modifications are included in Skyline's list of available isotope modifications. However, if you choose to use the wildcard modifications, the ProteomeXchange data validation process will automatically determine the correct Unimod modifications based on the amino acids in the peptides where the modification is applied. The matches found will be displayed in the
Modifications table of the validation results page.










