Biognosys PQ500 Introduction
This tutorial will show you how to create a targeted MS2 assay that uses heavy standards for absolute quantitation. The Biognosys PQ500 standard is used as the source of heavy standards. We used the Vanquish Neo LC, ES906A column and a trap-and-elute injection scheme with a 60 SPD method and a 100 SPD method. The gradients have been designed so that compounds elute over a large portion of the experiment spans.

Setting up the Skyline Document
Pierce retention time calibration mixture (PRTC) is used here to create an indexed retention time (iRT) calculator. Along with a spectral library, the iRT calculator will aid Skyline in picking the correct LC peaks in the steps that follow. See the Skyline iRT tutorial for more details. Here we will use an iRT calculator created with Koina. After setting up the LC and column, we run unscheduled PRTC injections to ensure that the LC and MS system is stable. The method file 60SPD_PRTC_Unscheduled.meth can be used for this. The prtc_unscheduled.csv file could be used to import into a tMSn table if making a method from scratch. We like to use Auto QC with Panorama to store all our files, and to automatically upload and visualize QC data.
Now we will create a Skyline document for analyzing PQ500 heavy labeled peptides. Biognosys supplies a transition list with intensities and iRT values that can be used to create a spectral library and iRT calculator. We'll show you though also how you can use use Koina integration with Skyline to create a spectral library and iRT library from a list of peptides sequences if you don't know anything about them. We also tend to like to use Koina spectral libraries even if supplied lists of transitions, because we will be using PRM Conductor to automatically filter the transitions. At the end of this section, we’ll be ready to perform unscheduled PRM for the PQ500 heavy-labeled peptide standards.
Transition Settings
Open up Skyline Daily and create a new document. Save the document as Step 1. Setup Skyline Documents/pq500_60spd_neat_multireplicate.sky.
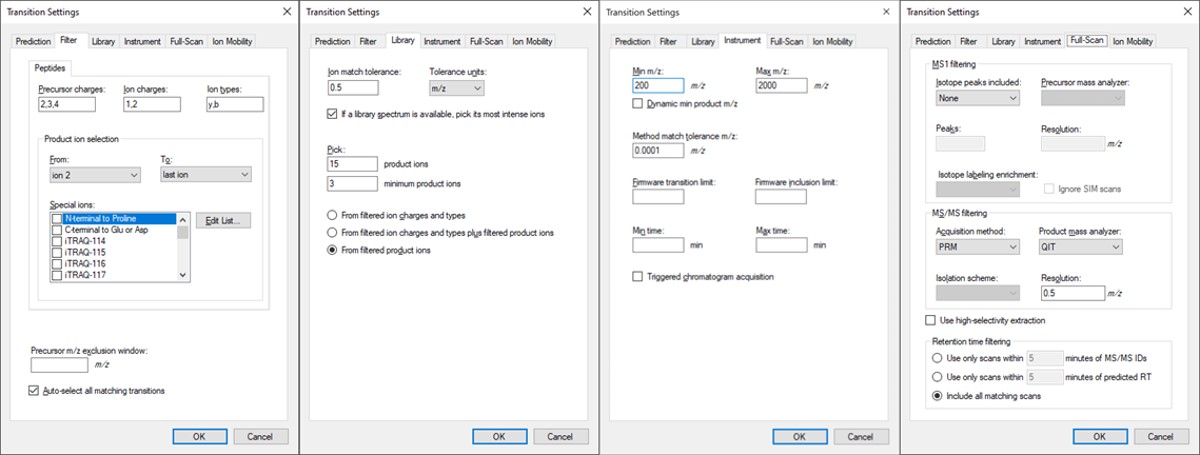
- Open Settings / Transition Settings / Filter. Set Precursor charges to ‘2,3,4’, Ion Charges to ‘1,2’, and Ion types to ‘y,b’. In the Product ion selection section, select ‘From ion 2’ to ‘last ion’. Un-select N-terminal to Proline and keep “Auto-select all matching transitions” checked.
- In the Library tab, set Ion match tolerance 0.5, check the box “If a library spectrum…”, set Pick 15 product ions with minimum 3 product ions. 15 is a large number, but we will refine the transitions later with the PRM Conductor tool. Select “From filtered product ions”.
- In the Instrument tab, set Min m/z 200, and Max m/z 2000. Set the Method match tolerance m/z to 0.0001. This helps Skyline to differentiate between precursors that have very close m/z. As we’ll see later, there are sometimes still peptides with different sequences but the same exact m/z.
- In the Full-Scan tab, set MS1 filtering / Isotope peaks included to None. If there are any precursor transitions, PRM Conductor will think that the document is in DDA mode, and will not be full featured. Set MS/MS filtering / Acquisition Method to PRM, Product mass analyzer to QIT, and Resolution to 0.5 m/z. Set Retention time filtering to Include all matching scans. Press Okay to close the Transition Settings.
Peptide Settings
Open Settings / Peptide Settings.
- In the Library tab, uncheck or remove any libraries that are there, for simplicity's sake.
- In the Modifications tab, make sure that Carbamidomethyl (Cysteine) and Oxidation (Methionine) modifications are enabled. The C-term R and C-term K isotope modifications should be enabled. Isotope label type and Internal standard type should be set to heavy.

iRT Calculator from PRTC
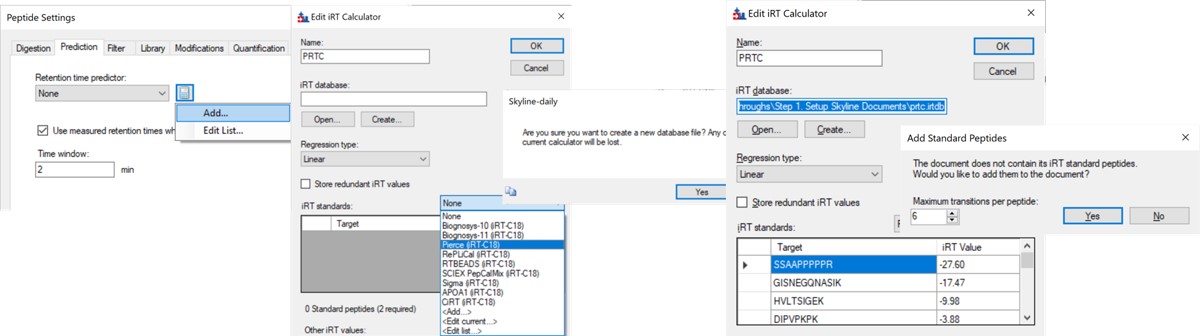
- In the Prediction tab, select the calculator icon and press Add. Add a name like PRTC. In the iRT Standards drop-down menu, select Pierce (iRT-C18). Press the Create button, and select Yes when asked if you want to create a new database file. Give the file a name like prtc.irtdb. Press Okay.
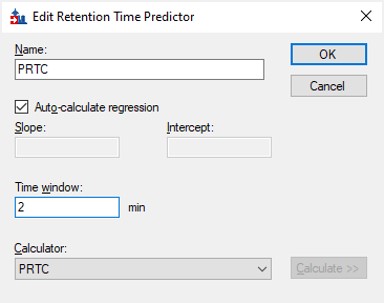
- Go back into the Peptide Settings / Prediction tab, click the dropdown arrow and select Edit current. Set the window to 2 minutes, and press Okay.
Importing the Transition List
Use File / Import / Transition List and select the file Step 1. Setup Skyline Documents/biognosis_pq500_transition_list.csv. A dialog opens that shows the mapping of the file headers to Skyline variable names. Press Okay to continue on. A new dialog prompts us that 624 transitions are not recognized. These are water losses that we don't necessarily need. We could define water loss transitions in the Settings tabs if we really wanted them. Press Okay twice to exit the iRT calculator dialogs. A new dialog will ask if you want to add the Standard Peptides, choose 6 transitions and press Yes.
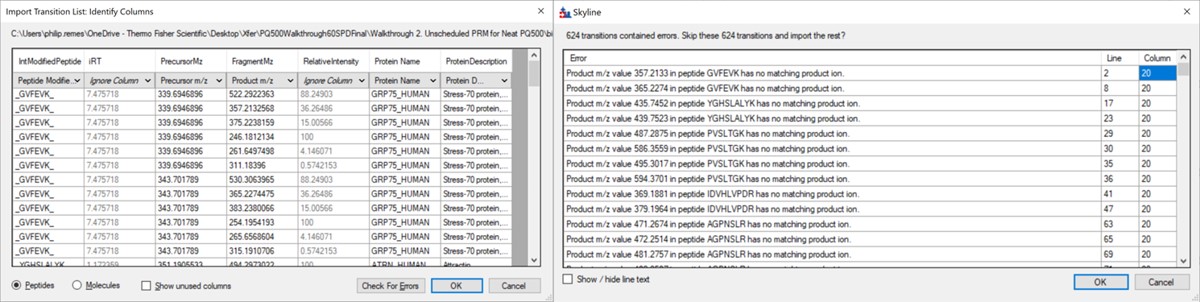
Another dialog appears, asking if we want to make an iRT calculator. The Biognosys values are presumably based on experiment, and are slightly more accurate than the in silico predicted iRT values from Koina, so Click Create. You'll be asked if you want to create a spectral library from the intensities in the transition list. Feel free to press Create if you want, but we will press Skip and use Koina to predict the intensities next. The Skyline document will update and in the bottom right border will be displayed 579 prot, 818 pep, 1622 prec, 9020 tran.

Koina
Now we’ll generate a spectral library with Koina.
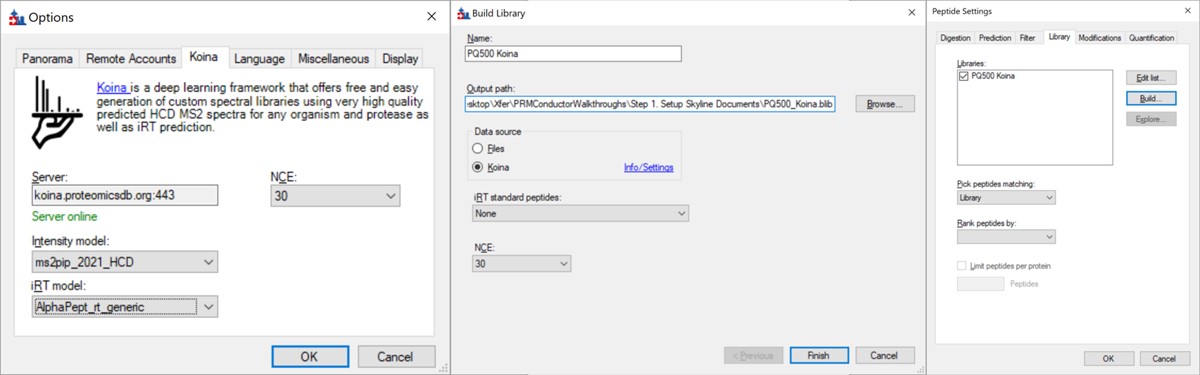
- Open Tools / Options and go to the Koina tab. Set the intensity model to ms2pip_2021_HCD, the iRT model to AlphaPept_rt_generic, and NCE to 30. Note that our original assay was created when this option tab said Prosit, but it had changed in a new Skyline Daily release by the time that this walkthrough was created. Press Okay.
- Go to Settings / Peptide Settings / Library / Create and select Data source Koina, NCE 30, and set a name and output path for the library. Press Finish. Make sure the library is selected in Peptide Settings Library and press Okay. If everything works correctly and you have internet access, your computer will communicate with the Koina server and return the spectral library to you, and the peptides in the Skyline tree will receive little spectra images to the left of the sequences, like below.
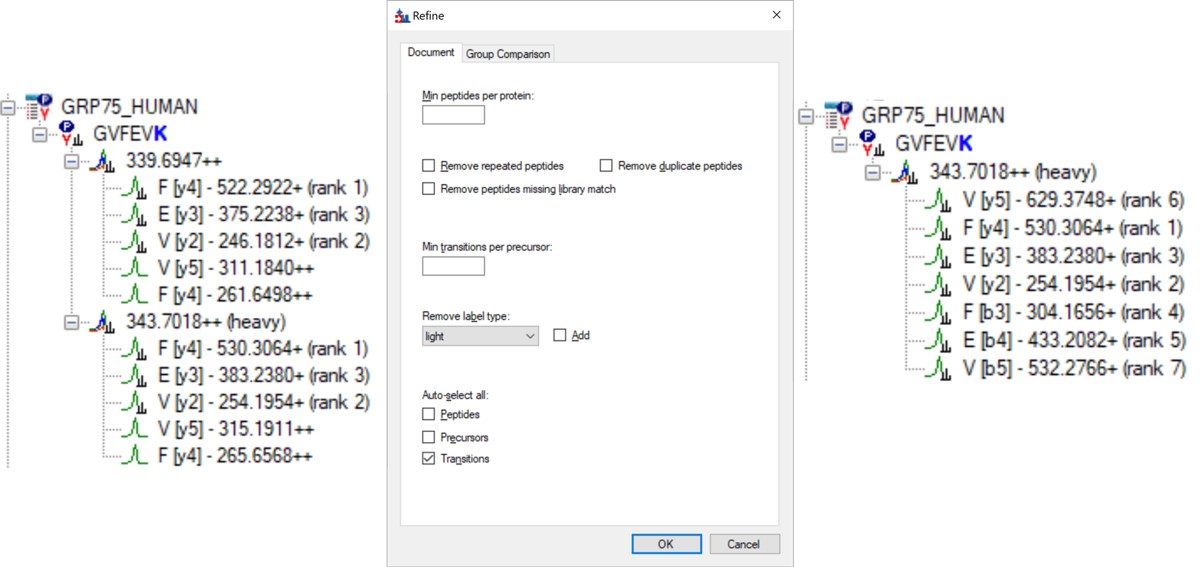
- Open Refine / Advanced and select Remove label type light, and click the box for Auto-select all Transitions, and press Okay. If you expand a peptide, like the first one, GVFEVK, it will have up to 15 transitions, instead of the up to 6 in the Biognosys transition file. Save the Skyline document. This is the end of Step 1: Setting up the Skyline Document.