Table of Contents |
guest 2024-07-04 |
Absolute Quantitation - PQ500
Neat Unscheduled Runs
Plasma Wide Window Scheduled
Plasma Narrow Windows Scheduled
Label Free - E. coli
Validation with Subset Replicates
Analysis of Final Assay Replicates
Introduction
Introduction
PRM Conductor is a software program that plugs into the Skyline External Tool ecosystem. The purpose of PRM Conductor is to aid in the creation of parallel reaction monitoring (PRM) mass spectrometry methods. The basic functions of the program are as follows:
- Receive DIA or PRM data
- Filter transitions with a set of filters
- Filter precursors that have at least N good transitions
- Visualize the scheduling of one or more PRM assays with particular acquisition parameters
- Export an instrument method for the new assay(s)
- Support the setup of the Adaptive RT algorithm for real-time chromatogram alignment
Location of the Walkthrough Materials
All the documents needed to perform the walkthroughs can be found by clicking the Raw Data tab on the top right hand side of this page.

Example Scenario
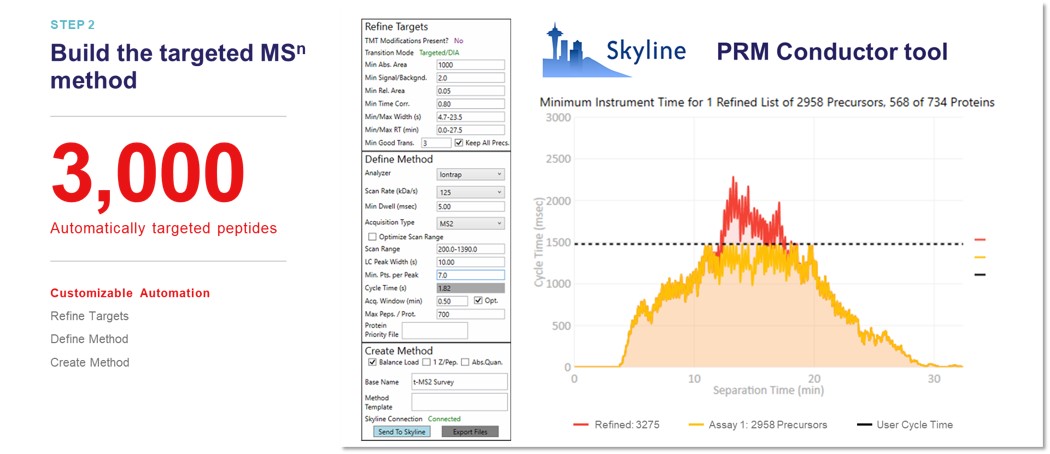
PRM Conductor is launched from Tools / Thermo / PRM Conductor. In one test case, we imported DIA data containing more than 5000 unique peptide identifications.

After applying the transition filters listed in the Refine Targets section, there were about 3000 'good' precursors left. The user can alter parameters in the Define Method, such as the Analyzer type, the minimum points per LC peak, and the scheduled acquisition window, and see how the 'good' precursors fit into a scheduled assay. The yellow trace shows precursors that can be acquired in less than the cycle time, while the red trace shows those precursors that can't be acquired. The user can then export a method with the Create Method section, where they can specify an instrument method template with LC details, which will be used to create a new method with the acquisition settings and precursor list filled in.
Walk-throughs
Several walk-throughs have been created to teach users how to make targeted methods with PRM Conductor. These methods generally fall into two categories at present; the absolute quantitation category, where there are heavy standards created for each endogenous peptide to be monitored, and the label-free category, where the assay is created directly from peptide search results, and there are no heavy standards.

Available Walk-throughs
- Absolute Quantitation
- PQ500
- Label Free
- E. coli in HeLa
Absolute Quantitation - PQ500
Biognosys PQ500 Introduction
This tutorial will show you how to create a targeted MS2 assay that uses heavy standards for absolute quantitation. The Biognosys PQ500 standard is used as the source of heavy standards. We used the Vanquish Neo LC, ES906A column and a trap-and-elute injection scheme with a 60 SPD method and a 100 SPD method. The gradients have been designed so that compounds elute over a large portion of the experiment spans.

Setting up the Skyline Document
Pierce retention time calibration mixture (PRTC) is used here to create an indexed retention time (iRT) calculator. Along with a spectral library, the iRT calculator will aid Skyline in picking the correct LC peaks in the steps that follow. See the Skyline iRT tutorial for more details. Here we will use an iRT calculator created with Koina. After setting up the LC and column, we run unscheduled PRTC injections to ensure that the LC and MS system is stable. The method file 60SPD_PRTC_Unscheduled.meth can be used for this. The prtc_unscheduled.csv file could be used to import into a tMSn table if making a method from scratch. We like to use Auto QC with Panorama to store all our files, and to automatically upload and visualize QC data.
Now we will create a Skyline document for analyzing PQ500 heavy labeled peptides. Biognosys supplies a transition list with intensities and iRT values that can be used to create a spectral library and iRT calculator. We'll show you though also how you can use use Koina integration with Skyline to create a spectral library and iRT library from a list of peptides sequences if you don't know anything about them. We also tend to like to use Koina spectral libraries even if supplied lists of transitions, because we will be using PRM Conductor to automatically filter the transitions. At the end of this section, we’ll be ready to perform unscheduled PRM for the PQ500 heavy-labeled peptide standards.
Transition Settings
Open up Skyline Daily and create a new document. Save the document as Step 1. Setup Skyline Documents/pq500_60spd_neat_multireplicate.sky.
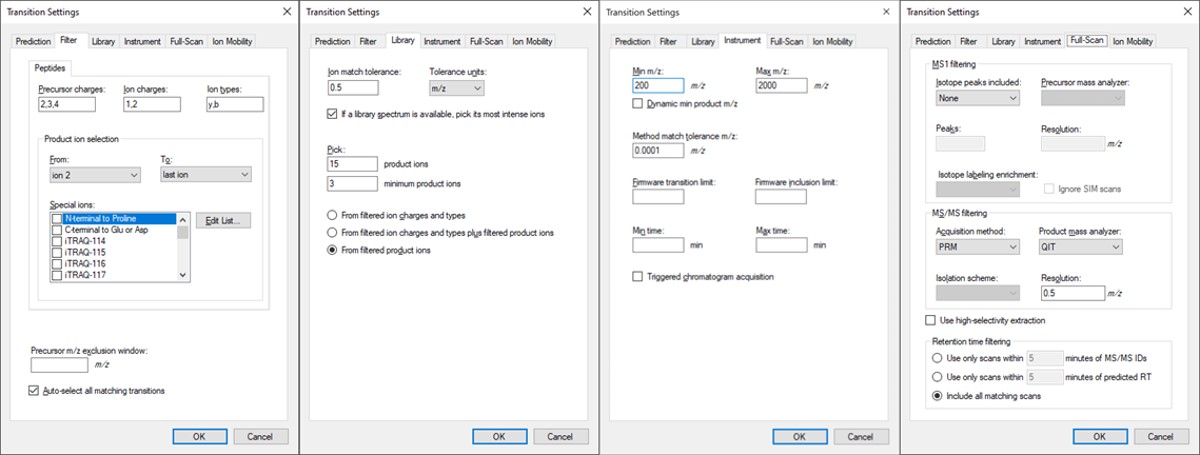
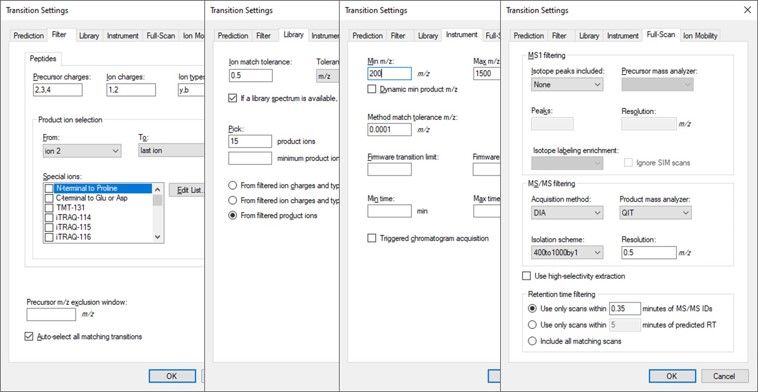
- Open Settings / Transition Settings / Filter. Set Precursor charges to ‘2,3,4’, Ion Charges to ‘1,2’, and Ion types to ‘y,b’. In the Product ion selection section, select ‘From ion 2’ to ‘last ion’. Un-select N-terminal to Proline and keep “Auto-select all matching transitions” checked.
- In the Library tab, set Ion match tolerance 0.5, check the box “If a library spectrum…”, set Pick 15 product ions with minimum 3 product ions. 15 is a large number, but we will refine the transitions later with the PRM Conductor tool. Select “From filtered product ions”.
- In the Instrument tab, set Min m/z 200, and Max m/z 2000. Set the Method match tolerance m/z to 0.0001. This helps Skyline to differentiate between precursors that have very close m/z. As we’ll see later, there are sometimes still peptides with different sequences but the same exact m/z.
- In the Full-Scan tab, set MS1 filtering / Isotope peaks included to None. If there are any precursor transitions, PRM Conductor will think that the document is in DDA mode, and will not be full featured. Set MS/MS filtering / Acquisition Method to PRM, Product mass analyzer to QIT, and Resolution to 0.5 m/z. Set Retention time filtering to Include all matching scans. Press Okay to close the Transition Settings.
Peptide Settings
Open Settings / Peptide Settings.
- In the Library tab, uncheck or remove any libraries that are there, for simplicity's sake.
- In the Modifications tab, make sure that Carbamidomethyl (Cysteine) and Oxidation (Methionine) modifications are enabled. The C-term R and C-term K isotope modifications should be enabled. Isotope label type and Internal standard type should be set to heavy.

iRT Calculator from PRTC
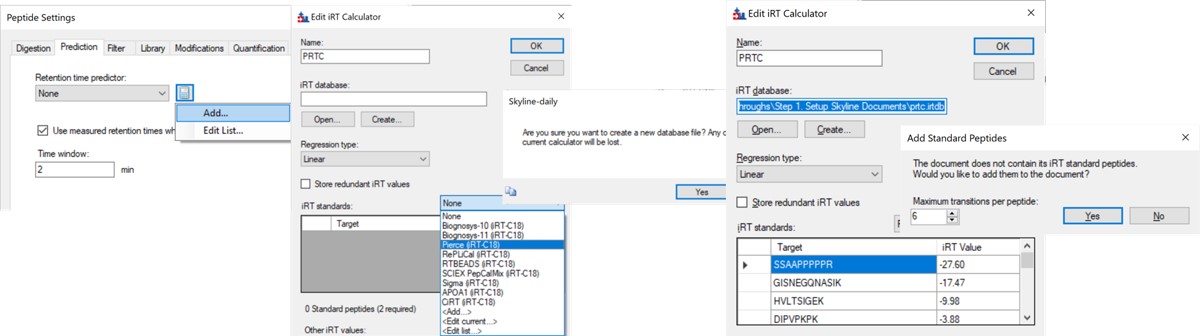
- In the Prediction tab, select the calculator icon and press Add. Add a name like PRTC. In the iRT Standards drop-down menu, select Pierce (iRT-C18). Press the Create button, and select Yes when asked if you want to create a new database file. Give the file a name like prtc.irtdb. Press Okay.
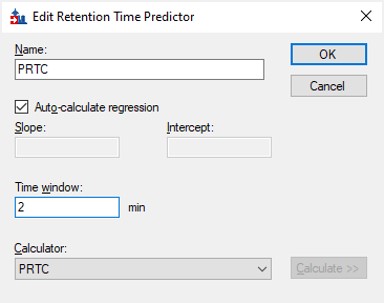
- Go back into the Peptide Settings / Prediction tab, click the dropdown arrow and select Edit current. Set the window to 2 minutes, and press Okay.
Importing the Transition List
Use File / Import / Transition List and select the file Step 1. Setup Skyline Documents/biognosis_pq500_transition_list.csv. A dialog opens that shows the mapping of the file headers to Skyline variable names. Press Okay to continue on. A new dialog prompts us that 624 transitions are not recognized. These are water losses that we don't necessarily need. We could define water loss transitions in the Settings tabs if we really wanted them. Press Okay twice to exit the iRT calculator dialogs. A new dialog will ask if you want to add the Standard Peptides, choose 6 transitions and press Yes.
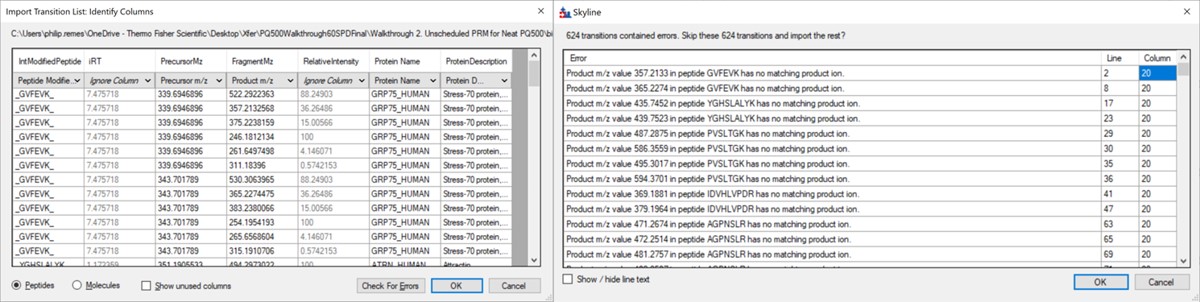
Another dialog appears, asking if we want to make an iRT calculator. The Biognosys values are presumably based on experiment, and are slightly more accurate than the in silico predicted iRT values from Koina, so Click Create. You'll be asked if you want to create a spectral library from the intensities in the transition list. Feel free to press Create if you want, but we will press Skip and use Koina to predict the intensities next. The Skyline document will update and in the bottom right border will be displayed 579 prot, 818 pep, 1622 prec, 9020 tran.

Koina
Now we’ll generate a spectral library with Koina.
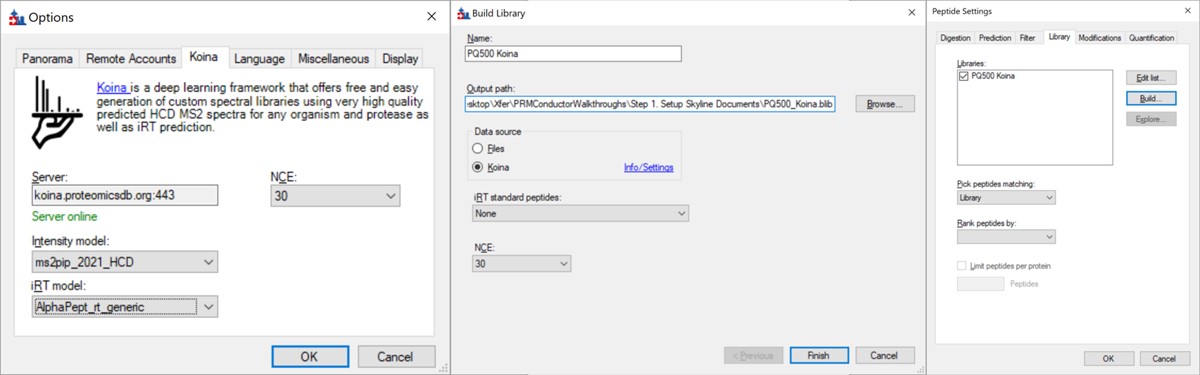
- Open Tools / Options and go to the Koina tab. Set the intensity model to ms2pip_2021_HCD, the iRT model to AlphaPept_rt_generic, and NCE to 30. Note that our original assay was created when this option tab said Prosit, but it had changed in a new Skyline Daily release by the time that this walkthrough was created. Press Okay.
- Go to Settings / Peptide Settings / Library / Create and select Data source Koina, NCE 30, and set a name and output path for the library. Press Finish. Make sure the library is selected in Peptide Settings Library and press Okay. If everything works correctly and you have internet access, your computer will communicate with the Koina server and return the spectral library to you, and the peptides in the Skyline tree will receive little spectra images to the left of the sequences, like below.
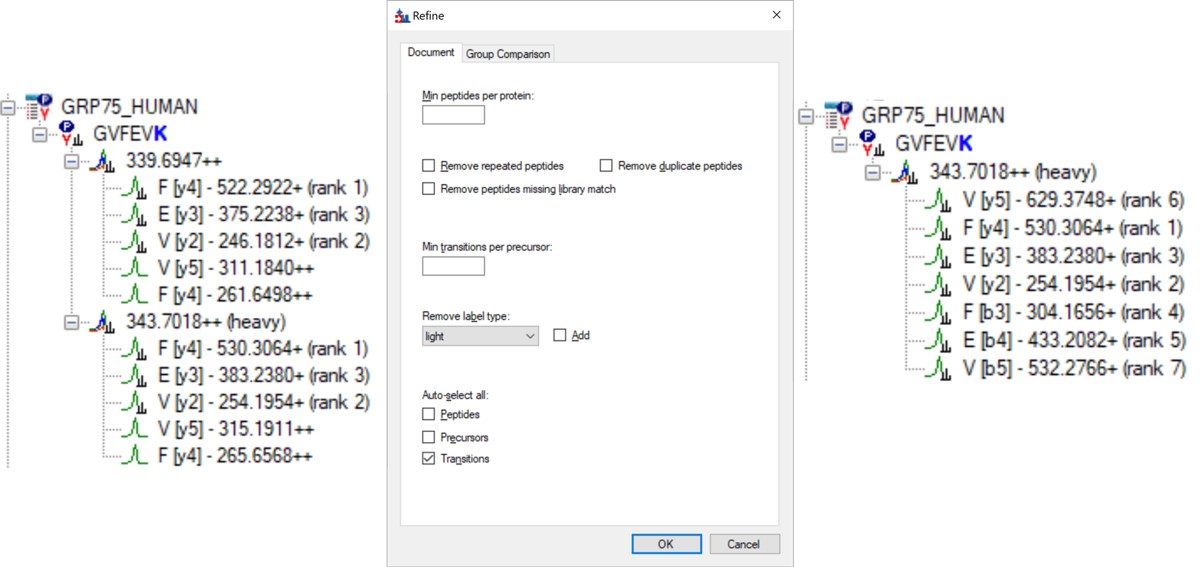
- Open Refine / Advanced and select Remove label type light, and click the box for Auto-select all Transitions, and press Okay. If you expand a peptide, like the first one, GVFEVK, it will have up to 15 transitions, instead of the up to 6 in the Biognosys transition file. Save the Skyline document. This is the end of Step 1: Setting up the Skyline Document.
Neat Unscheduled Runs
- Unscheduled PRM for Neat Heavy PQ500 with Multiple Replicates
Unscheduled PRM for Neat Heavy PQ500 with Multiple Replicates
In this step, we’ll use Skyline to create a set of unscheduled PRM methods for the 804 PQ500 peptides. At the end of this step, we will have created 10 Unscheduled PRM methods for both 60 and 100 SPD, acquired data for them, loaded the results into Skyline, and assessed the results. We’ll be ready to look at our standards spiked into matrix in the next step.
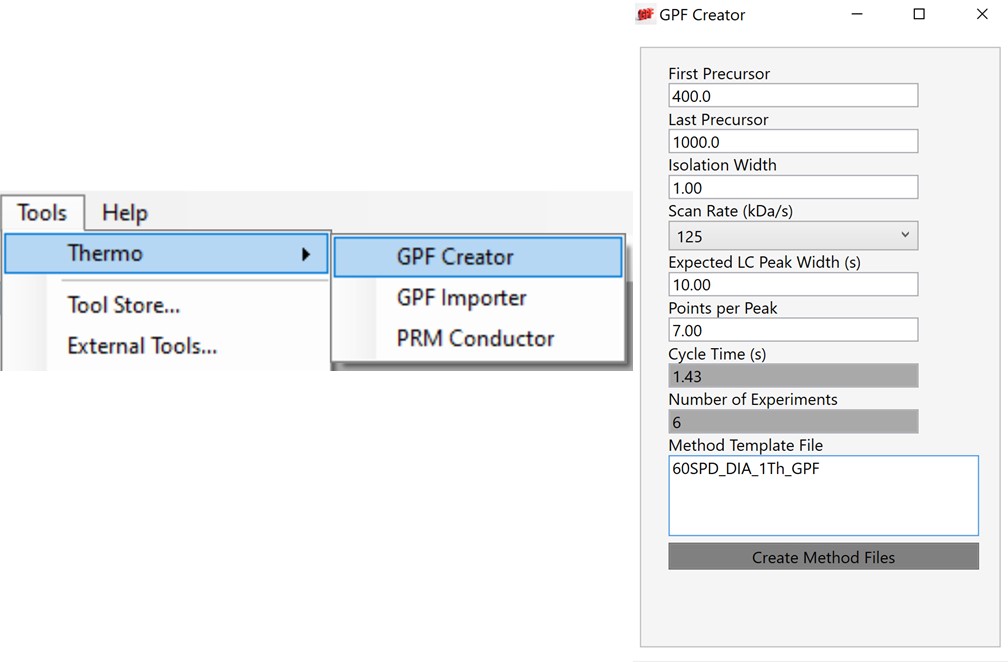
Alternative DIA GPF Acquisition
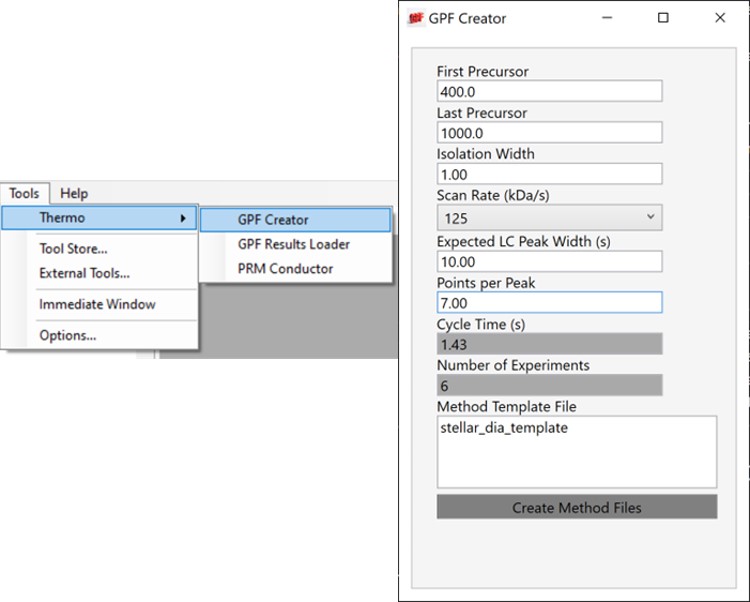
Alternatively, especially as the number of heavy peptides increases, one could opt to use data independent acquisition (DIA) of the neat, heavy standards to find their retention times. One would simply find the smallest and largest m/z of the peptides in question and use the Thermo method editor to create a DIA method. For example, we have had success in some neat standard cases using a single injection with 4 Th isolation width. However multiple gas-phase fractions (GPF) could be acquired with narrower isolation widths, as in the technique we use for identification of unknowns. We included a little helper application in our Thermo suite of external tools called GPF creator that spawns GPF instrument methods. Given a set of parameters, namely a precursor m/z range and a Stellar DIA method template, it will create a cloned set of methods with the appropriate Precursor m/z range filled in. In the case below with Precursor m/z range 400-1000 and 6 experiments, methods would be created for the ranges 400-500, 500-600, all the way to 900-1000. The resulting .raw files could be used in the much the same way that we’ll use the unscheduled PRM data files in the coming steps, only that we would have to configure the Skyline Transition / Full Scan / Acquisition to DIA with the appropriate window scheme (Ex. 400 to 1000 by 1 with Window Optimization On). As it is, we continue on, using the Unscheduled PRM technique.
Creating Unscheduled PRM Methods
The newest Skyline release supports Stellar for exporting isolation lists and whole methods, which is convenient as it saves the step of importing isolation lists for each of the methods. However, for completeness we'll also describe how to use the isolation list dialog with manual import into method files.
Isolation List with Manual Import to Instrument Method Files
- Open the dialog using File / Export / Isolation List. Set Instrument type Thermo Stellar, select Multiple methods, Max precursors per sample injection to 100, Method type Standard. Note that the max value of 100 is approximate. A smaller number could be required for more narrow peaks or shorter gradients. Press Okay.
- A Save-As dialog will open. Use the name pq500_neat_unscheduled in the folder Step 2. Neat Unscheduled Multireplicates. In a moment, 10 .csv files with the suffix _0001 to _0010 will appear in the folder. An important aspect of the iRT workflow is that Skyline included the PRTC compounds in each of the 10 files. This allows Skyline to calculate the relative positions of the rest of the peaks, which will eventually be added to our iRT library.
- Open the Stellar method editor and open the file Step 2. Neat Unscheduled Multireplicates/pq500_60spd_neat_multireplicate.meth. In the tMSn experiment in the bottom right you can find the Mass List table and press the Import button to load the first of the created isolation lists, pq500_neat_unscheduled_0001.csv. Use File / Save-As to save the method as pq500_neat_unscheduled_0001.meth. Do this for each of the other 9 isolation lists, creating a total of 10 methods. At this point you would create an Xcalibur sequence with these 10 methods, injecting an appropriate amount of neat standard (ex 10-100 fmol for a 1 ul/min, 24 min assay), and creating 10 raw files. We have performed this experiment for both 60 and 100 SPD methods, and put the resulting .raw files in the folder Step 2. Neat Unscheduled Multireplicates/Raw.

Skyline Method Export
The more convenient way to create the unscheduled replicates is to use the Skyline File/Export/Method functionality. In the Export Method dialog, select Instrument type Thermo Stellar. Select Multiple methods, with Max precursors per sample injection 100. This is a ballpark number that has given enough points per peak for identification purposes for neat standards for a variety of experiment lengths. Click the Browse button and choose the pq500_60spd_neat_multireplicate.meth file in the Step 2. Neat Unscheduled Multireplicates folder. Use a name like pq500_60spd_neat_multireplicates and press Save, and Skyline will present a progress dialog. When it finishes, 10 new methods will be created with suffix _0001 through _0010, as shown below.
Loading Unscheduled PRM Data
- Take the .sky file that we have created, Step 1. Setup Skyline Documents/pq500_60spd_neat_multireplicate.sky and use File / Save As to save a copy with the name Step 2. Neat Unscheduled Multireplicates/pq500_60spd_neat_multireplicate_results.sky. If you want to work along with the 100 SPD method as well, you can save another version of the file with the corresponding name.
- Use File / Import / Results and select Add one new replicate. Give it the name NeatMultiReplicate. Press Okay, and then select all the raw files in the folder Step 2. Neat Unscheduled Multireplicates\Raw\NeatUnscheduled60SPD and press Open. Skyline will load the results. When they are finished loading, Save the Skyline document. Do the same for the 100 SPD .raw files in their .sky file.

- Now we will inspect the results to see if any peaks have been chosen incorrectly. Use File / Save As to save the Skyline document as Step 2. Neat Unscheduled Multireplicates/pq500_60spd_neat_multireplicate_results_refined.sky (and one for the 100 SPD file).
- Use View / Retention Times / Regression / Score to Run. Right click the plot and select Plot / Residuals. Right click the plot and make sure that Calculator is set to the calculator that we created, PRTC.
- Right click on the plot and select Set Threshold and enter 0.99. If the plot does not update to have some of the blue dots turn pink, then right click the plot and select Plot / Correlation, and then switch back to Plot / Residuals.
- Use View / Peak Areas / Replicates Comparison and setup the Skyline document something like in the figure below.
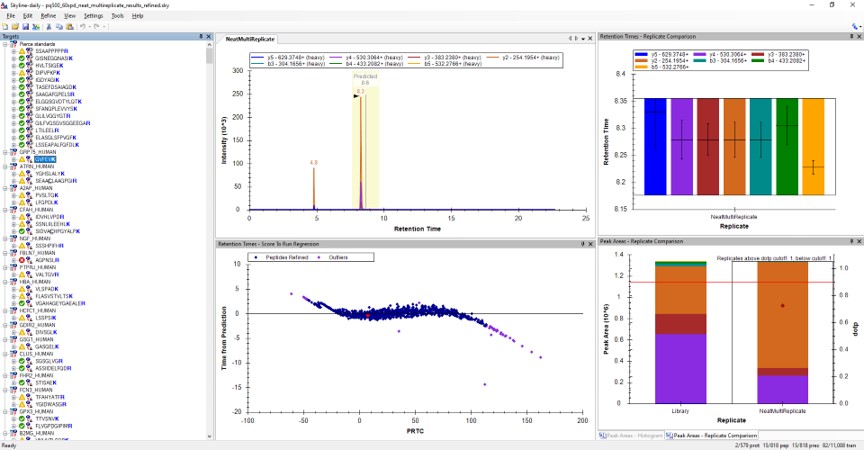
Reviewing the Unscheduled PRM Results
Using Score-to-Run RT Outliers
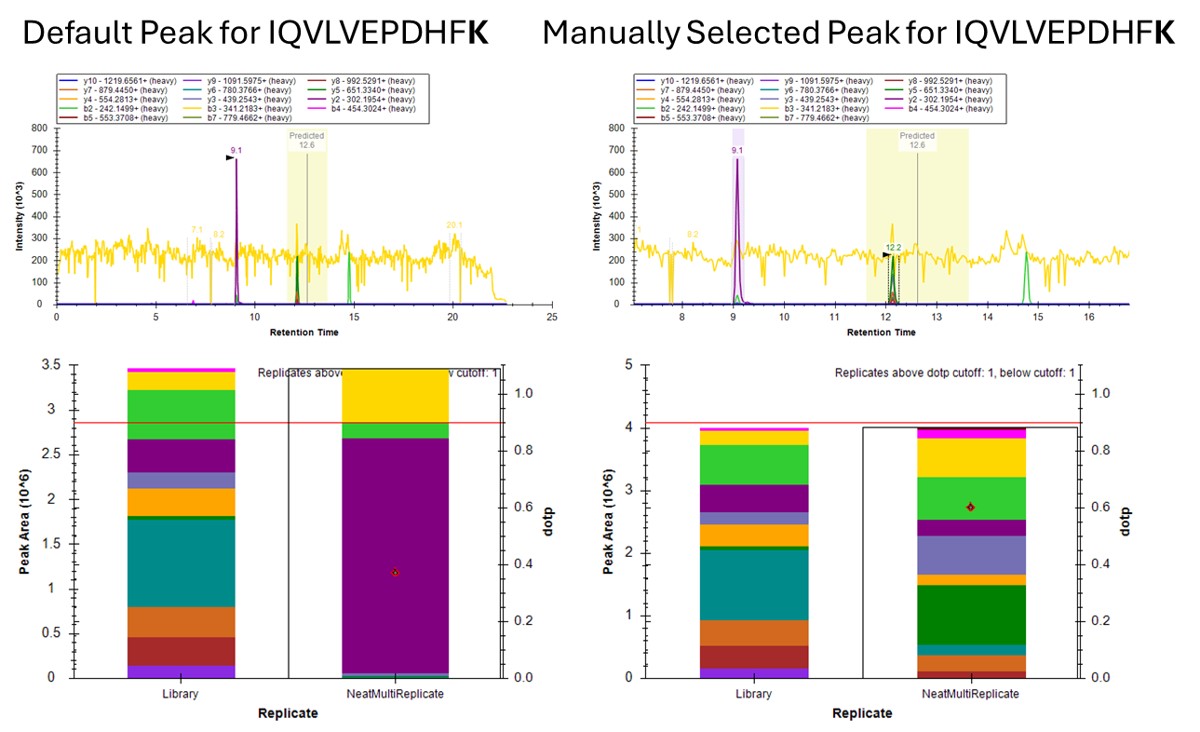
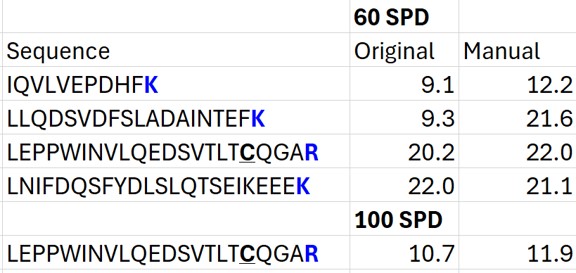
- The Residual Score-to-Run plot is very useful for this step to flag any potential missed peaks. Here we want to click on any of the pink "outlier" dots and inspect them. For example the IQVLVEPDHFK was picked at 9.1 minutes, but the peak in the predicted window at 12.6 has a better library dot product match. Also based on the 100 SPD data, we think that the LLQDSVDFSLADAINTEFK is probably the low intensity peak at 21.6 minutes and not the larger peak at 9.3 minutes that is far away from the predicted RT.
Using Dot Products
-
Another way to find picked peak issues like this is to view a Document grid report that has the dot product scores. Use View / Document Grid (or Alt + 3) to bring up the Document grid, and dock it in the same window as the Retention Time Score-to-Run.
- If you ever can't dock a window where you want it, dock the window in some place that Skyline lets you, and then you will be able to dock it where you originally wanted.
-
Click the Precursor Report, then Customize Report to bring up a dialog menu. Erase columns from the right hand side and then click the binoculars and type 'Dot', and press Find Next until you find Library Dot Product. Select this column to add it to the right hand side, and then press okay.
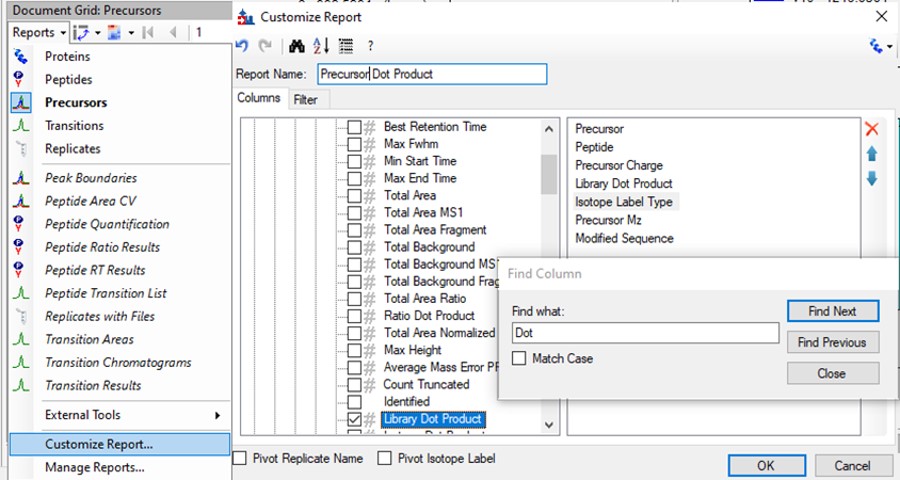
- Now right click on the Library Dot Product column and select Sort Ascending. You can click through on the worse dot product scores to inspect them. Even the ones with poor dot product scores are actually pretty ambiguously belonging to a single precursor at the correct retention time.
Using Peak Picking Models
A final way that can be useful for inspecting this kind of result is to compare the results from the two Skyline peak picking models available at this time. Although in the present case there is not much use for this technique, we'll demonstrate it now.
- Select Refine / Reintegrate. In the Peak scoring model dropdown box, select Add. In the dialog that shows up, keep mProphet as the model, and select Training / Use second best peaks. Using decoys is the method used when you have DIA data and have added Decoy peptides to the document. For targeted methods, the second best peak for the peptide serves as the null distribution for training the peak model. Click Train Model. In the feature scores there are several scores that are not available because there are no light peptides in the document. Scroll down the see the enabled features. Several features are red because they decreased the classification accuracy. Unselect those features and press Train Model again, and click Okay to exit the Edit Peak Scoring Model dialog.

- In the Reintegrate dialog, Add another peak scoring model. This time select the Default model and press Train Model. Then press Okay, and press Okay again in the Reintegrate dialog. This should give us the original peaks selected when the data were imported.

- Go to Refine / Compare Peak Scoring. Use the Add button to add the two peak models that we just made. Go to the Score Comparison tab, select the Default model for the Compare dropdown and the mProphet model for the "with" dropdown. Click the Show conficts only button. Of the 8 conflictin peaks, the greatest ambiguity is probably the first one, LLGNVLVCVLAR. Here there is a very nice peptide peak at 20.9 minutes, but the one at 20.5 although it is further from the predicted RT, has a better dot product. We selected the 20.9 peak, but potentially this is misassigned. We made the same assignment in the 100 SPD method.

Summary of Changes made to Default Skyline Peak Picking
The figure below summarizes the manual changes we made to the Skyline default peak picking. Note that at the time that we first did the study, Skyline could connect to the Prosit server for library generation. By the time this tutorial was written, Skyline was using something called Koina to do the in silico predictions. While the Prosit models are in theory supported, there was an issue with using them. Therefore there could be some small differences. Note that the most conservative approach would be to search the unscheduled PRM data against a PQ500 .fasta file with static R and C heavy modifications, and only select those peptides that passed some threshold FDR value.
Saving a New, Empirical iRT Calculator
- Now we will update the iRT library so that future experiments can benefit from this curated peak picking. Use Settings / Peptide Settings / Prediction, and click the calculator button to Edit Current. In the dialog that opens, click Add in the bottom right corner and add results.., and an Add iRT Peptides dialog announces that 804 peptide iRT values will be replaced. Click Okay. Click Yes when asked if the iRT Standard values should be recalibrated. Now in the iRT database line, use the name pq500_60spd.irtdb, and give it a new name, PQ500+PRTC 60 SPD, press Okay to exit.

- We've created a new iRT calculator, but have to set it as the active one. Click the calculator icon and Edit List. Press the Add button and then click the dropdown menu, and the new calculator will be there, select that one. Set the Time window to 2 minutes, then press Okay. Select the PQ500+PRTC 60 SPD calculator in the Peptide Settings / Prediction dialog and press Okay to exit to main Skyline window.
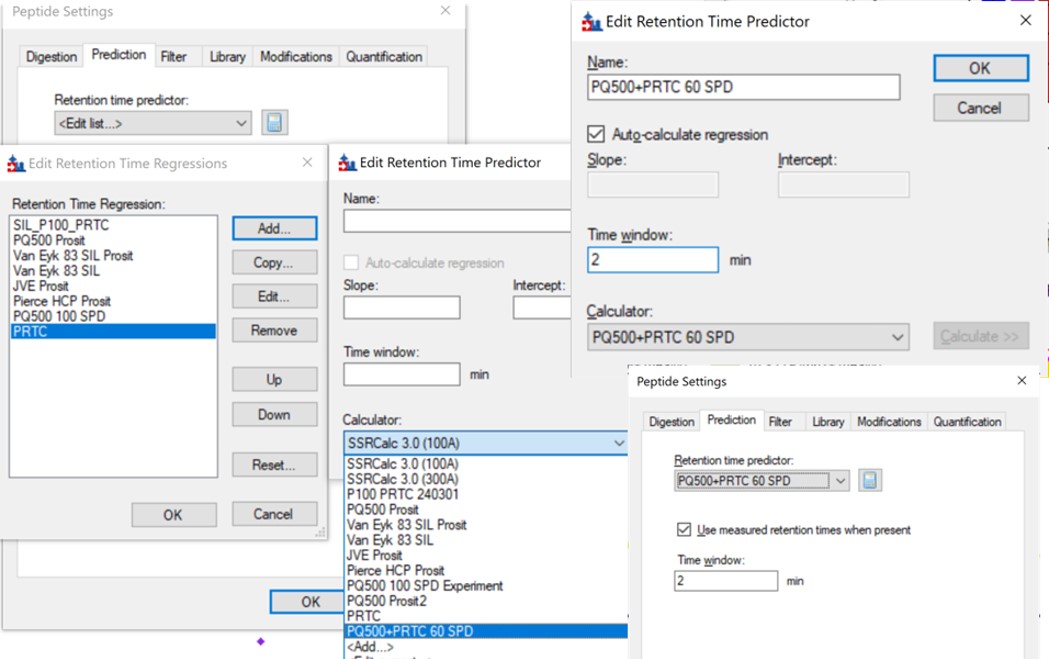
- In the score-to-run plot, right click, select Calculator, and choose the PQ500+PRTC 60 SPD. The plot will update to show an interesting pattern where the errors are all very close to zero. We have now created a new calculator that will make it easier for Skyline to pick peaks next time. We did the exact same set of iRT steps for the 100 SPD Skyline file, because using the new 60 SPD calculator for the 100 SPD does not work perfectly. In the 100 SPD we think that probably the LEPPWINVLQEDSVTLTCQGAR is the peak at 11.9 minutes and not the one at 10.7 minutes.
Filtering Transitions with PRM Conductor
This was a neat sample, but we can filter out the transitions that we don't need at this point, with the understanding that when we spike into plasma we may have to refine the transitions even further.
- Select Tools / Thermo / PRM Conductor. The tool opens up. We used the settings in the figure below, where importantly you set Min Good Trans to 5, press Enter, and also select the Keep All Precs box. Press the Send to Skyline button on the bottom left. Save the Skyline document and close the PRM Conductor.

- The number of transitions displayed in the bottom right hand of Skyline will have updated. With the settings we used, the number of transitions dropped from 11,088 to 7,564.

- Use File / Export / Spectral Library and save as Step 2. Neat Unscheduled Multireplicates/PQ500_Neat_60SPD_Refined. Use Settings / Peptide Settings / Library and select Edit List, then Add, and Browse for the file you just saved. Give the library the name PQ500 Neat 60 SPD. Press Okay, and in the Library tab, deselect the PQ500 Koina library and select the new library. Press Okay and save the Skyline document. Make a spectral library in the same way for the 100 SPD document. Close the PRM Conductor instances. We've reached the end of the second step, and are ready to acquire data in plasma.
Plasma Wide Window Scheduled
Scheduled PRM for Heavy PQ500 in Plasma with Wide Acquisition Windows
In this step we will create a wide-window PRM method to verify the RT locations of the PQ500 heavy peptides in plasma. Sometimes it can be the case that the RT’s of peptides will be much different when spiked into matrix compared to when analyzed neat. This is expected and likely due to the binding properties of the chromatography stationary phase, which depend on the concentration of analytes in the liquid phase in an equilibrium sometimes referred to as an isotherm. At the end of this section we will have a candidate final method that includes both heavy and light peptides, and that also includes Adaptive RT real-time chromatogram alignment.
Creating Wide Window Methods that Include Adaptive RT Acquisitions
-
Use File / Save As on our files from the last step, pq500_60spd_neat_multireplicate_results_refined.sky and pq500_100spd_neat_multireplicate_results_refined.sky and save in the folder Step 3. Plasma Heavy-Only Wide Window as pq500_60spd_plasma_multireplicate_results.sky and pq500_100spd_neat_multireplicate_results_refined.sky.
-
Use Tools / Thermo / PRM Conductor.
-
Update the settings as in the figure below. After changing any number value, be sure to press the Enter key on the keyboard. The prtc_priority.prot file is selected by double clicking the Protein Priority File text box. This is just a text file with the line “Pierce standards”, the protein name that Skyline gave to the iRT standards. The peptides from any proteins listed in this file (with Skyline's protein names, not accession numbers) are included in the assay, whether or not their transitions meet the requirements. If the Balance Load checkbox is not selected and there are multiple assays to export, each assay will contain the prioritized proteins, and Skyline will be able to use the iRT calculator for more robust peak picking.
-
Note that with the 1.8 minute acquisition window, the right-most plot in PRM Conductor tells us that the 818 precursors in the assay require up to almost 2500 milliseconds to be acquired, and as we have the Balance load box unchecked, they will be split into 2 assays. If Balance Load was checked, then we would create a single assay, for only the precursors that can be acquired in less than the Cycle Time.

-
Enter a suitable Base Name like PQ500_60SPD_Plasma_ToAlign. This reflects the fact that we are including acquisitions to perform Adaptive RT, but we are not actually adjusting our scheduling windows in real time. Our neat standards were not suitable for performing aligning in the complex plasma matrix background.
-
Double click the Method Template field and select the Step 3. Plasma Heavy-Only Wide Window/PQ500_60SPD_ToAlignTemplate.meth file. This file is standard targeted method for Stellar, with 3 experiments. The first is the Adaptive RT DIA experiment, which is being used to gather data for real-time alignment in future targeted methods. The second is a MS1 experiment, which isn't strictly needed, but enables the TIC Normalization feature in Skyline to be used, and can be helpful for diagnostic purposes. Removing it would save on computer disk space. The tMSn experiment can be simply the default tMSn experiment, where we ensure that Dynamic Time Scheduling is Off. If it were on, then PRM Conductor would try to embed alignment spectra from the current data set into the method. Here we leave it off.

-
Press the Export Button. PRM Conductor will open a progress bar and do some work to export a .sky file, and two .meth files with the names PQ500_60SPD_Plasma_ToAlign_0.sky and PQ500_60SPD_Plasma_ToAlign_1.sky. The new .sky file has our new transition list imported and sets the Acquisition mode to PRM. This can be useful especially when discovery data is acquired in a DIA mode, however in this case we want to still compare our neat PQ500 data with the spiked plasma data we’ll be collecting, so we’ll continue using our file pq500_60spd_plasma_multireplicate_results.sky.
-
Do the same thing for the 100 SPD method. Here we can create 3 methods if the LC Peak Width is set to 8. Change the Base Name and Method Template to the 100 SPD versions and press Export Files.
- The created methods in this step are as in the figure below. We are now ready to acquire data using these methods. We did this by spiking in PQ500 at the Biognosys recommended concentration into 300 ng of digested plasma that we got from Pierce. Pierce will offer this as a product for sale in the near future (this is written mid-2024).
Reviewing the Wide Window, Plasma, Scheduled PRM Results
-
Open Settings / Transition Settings and set the Retention time filtering option to Use only scans within 1 minutes of MS/MS IDs. You want to be careful with this filtering because if the RT shifts were greater than +/- 1 minute, some data could be missing. You can always use one number and then change it, and use Edit/Manage Results, and select the replicate and Reimport, to use a wider or narrower filter. In this case the IDs are coming from the spectral library that we created in the previous step. Alternatively one could use Use only scans with X minutes of predicted RT option. We need some kind of RT filtering of this sort to help Skyline differentiate between the iRT peptide SSAAPPPPPR and the PQ500 peptide FQASVATPR, which have the same exact m/z.
-
We acquired data for PQ500 spiked into 300 ng of plasma and put the resulting .raw files in the folder Step 3. Plasma Heavy-Only Wide Window\Raw. Load the results with File / Import Results / Add one replicate, use a name like PlasmaMultireplicate, and select the .raw files for the appropriate 60 or 100 SPD throughput. Skyline will load these data files as a single replicate.
-
Use View / Arrange Graphs / Row so that we can view the Neat and the Plasma replicates at the same time. Right click a chromatogram plot and use Auto Zoom X Axis / None, so that we are zoomed out as far as we can go.
-
Use View / Retention Times / Regression / Run-to-Run. Your Skyline document should look something like the figure below.
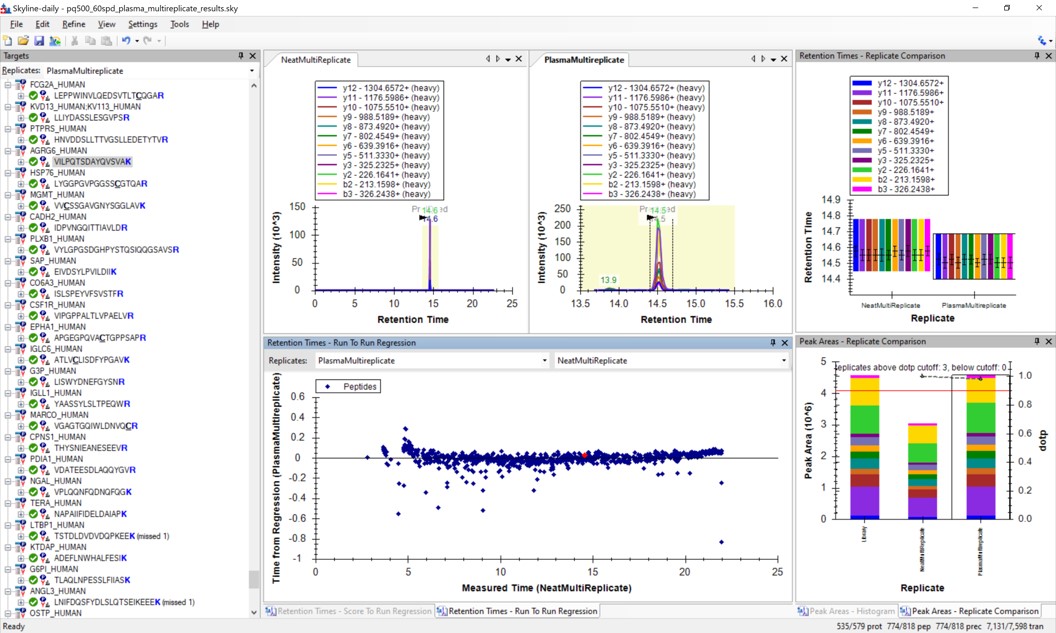
-
Use Save As to save a new version of this file in case we make any changes to the picked peaks. You can use pq500_60spd_plasma_multireplicate_results_refined.sky.
-
We can do the same steps as above. Make an mProphet and Default Peak picking models with Refine / Reintegrate. Then use Refine / Compare Peak Scoring, select the two new models with the Add button, then select the Score Comparison tab. Select the two models, and click Show conflicts only. We see two descrepancies, and changed the ELLALIQLER peptide from 20.0 to 20.2 minutes, which had a higher dot product and better predicted RT. We kept the default peak for the other peptide.
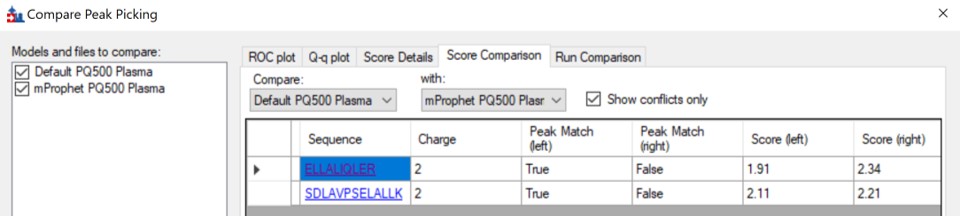
-
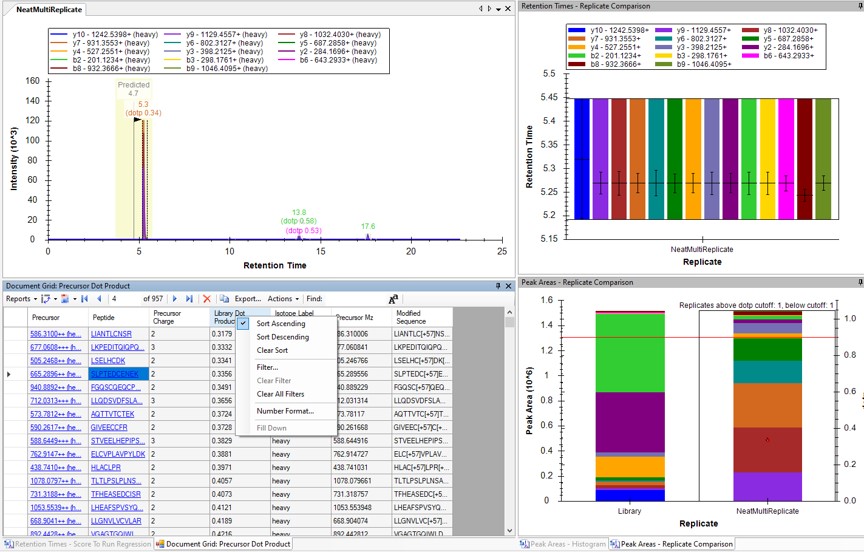
Click on the various outliers and make sure that the peak area plots are showing a good correspondance of the transitions with a high dot product.
-
Use a report with the Library Dot Product sorted from Low-to-High and investigate the worst cases. Even the lowest dot product cases look okay to us.
-
For the 100 SPD data, investigating the Plasma-to-Neat retention times has a similar patterns as for the 60 SPD. There is one case, the FQASVATPR peptide, that has the same exact m/z as the SSAAPPPPPR iRT peptide, which elutes at a similar RT. Reducing the Settings / Transition Settings / RT filtering time to +/- 0.5 minutes can separate these two peptides. We kept all the Skyline picked LC peaks for the 100 SPD document, and saved a new file pq500_100spd_plasma_multireplicate_results_refined.sky.
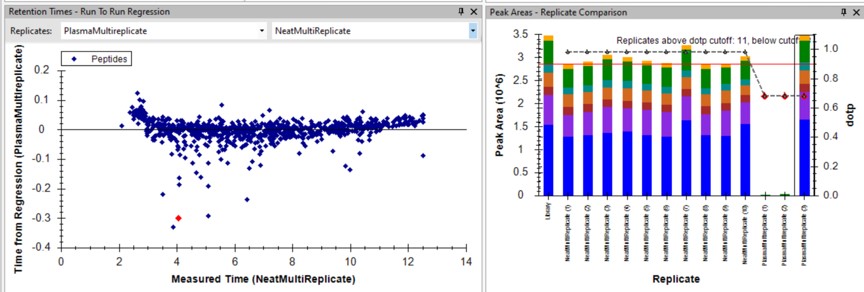
Creating a Final Method
-
Remove the NeatMultiReplicate using Edit / Manage Results. It's easy to forget to do this. We don’t want PRM Conductor to consider the neat peaks, which are already very clean. Save the Skyline file again.
-
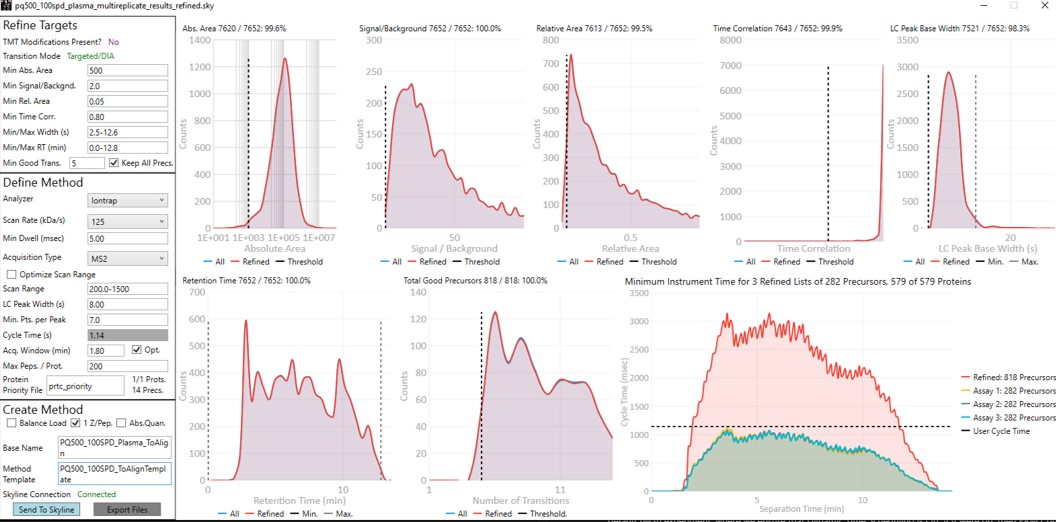
Launch PRM Conductor to clean up interferences and create a final method. Set Min. Good Trans. 5 and check Keep All Precs. Set Min Dwell 5 msec. Set LC Peak Width 11, Min. Pts. Per Peak 7, Acquisition Window 0.6 minutes, and check the Opt. box. This option increases the acquisition windows slightly, especially at the start of the experiment, without going over the user's Cycle Time. Select the prtc_priority.prot file, which in in this case just makes sure that those peptides can't get filtered. Check the Balance Load, 1 Z/prec., and Abs. Quan boxes. This last option instructs the Export command to include light targets for each of the heavy targets. Set a Base Name PQ500_60SPD_Align, and select the PQ500_60SPD_AlignTemplate.meth.
-
This template method is the same as the ToAlign version, only the Dynamic Time Scheduling is set to Adaptive RT. Now when PRM Conductor exports a method, it will compress the qualifying alignment acquisitions in the data and embed them into the created method.
-
We have a small issue here in that there more refined targets (red trace) than we can target. We have to trick PRM Conductor here and set the LC Peak Width to 20 so that all targets are exported, then in the created file change the LC Peak width back to 11 and points per peak to 7. In the future we'll allow the user to just export an "invalid" method.
-
Press Export Files to create the new instrument method.

- Open the PQ500_60SPD_Align.meth file, change the LC Peak width from 20 to 11, and the Points per Peak to 7. Note that the Adaptive RT Reference file has a file name that starts with Embedded, and the Mass List table has entries with heavy peptides having names ending in [+10] or [+8] followed by the corresponding light peptides.

-
Use the Send to Skyline to filter the remaining few poor transitions from these targets, and save the Skyline document state. Export a spectral library like we did before, giving it a name like PQ500_60SPD_Plasma. Configure this library in Settings / Peptide Settings / Library. This is the end of step 3.
-
For the 100 SPD case, from the pq500_100spd_plasma_multireplicate_results_refined.sky file, Launch PRM Conductor. Check the Optimize Scan Range box. This will produce targets with customized scan ranges for each target, significantly increasing the acquisition speed, at the cost of some injection time and sensitivity. Set an appropriate Base Name and select the PQ500_100SPD_AlignTemplate.meth for the Method Template. Use the same trick as for the 60 SPD, setting the LC peak width to 20 seconds and Export the method. Then open the method that is created and change the LC peak width back to 7 with 6 points per peak.
-
Press the Send to Skyline button. Export the spectral library and configure it in the Peptide Settings / Library tab. Save the pq500_100spd_plasma_multireplicate_results_refined.sky.

- We now have candidate final methods created for 60 SPD and 100 SPD, respectively PQ500_60SPD_Align.meth and PQ500_100SPD_Align.meth. We are ready to move to the next step and validate the assay.
Plasma Narrow Windows Scheduled
Scheduled PRM for Light-Heavy PQ500 in Plasma with Narrow Acquisition Windows
Analysis of the Heavy Peptides
In step 3 we created two candidate final methods for the 60 and 100 SPD assays. Take the file pq500_60spd_plasma_multireplicate_results_refined.sky and pq500_1000spd_plasma_multireplicate_results_refined.sky, and resave them in the folder Step 4. Plasma Light-Heavy Narrow Window, with names like pq500_60spd_plasma_final_replicates.sky and _pq500_100spd_plasma_final_replicates.sky. In this step we’ll analyze the results of the light/heavy methods created in Step 3. Of particular interest will be the histogram of coefficient of variance values for the peak areas.
-
Use File / Import Results / Add single-injection replicates in files and press Okay. Select the 10 files in Step 4. Plasma Light-Heavy Narrow Window\Raw\60SPDReplicates and press Open. Remove the common prefix and press Okay to load the results. Remove the PlasmaMultiReplicate with Edit / Manage Results and Save the document.
-
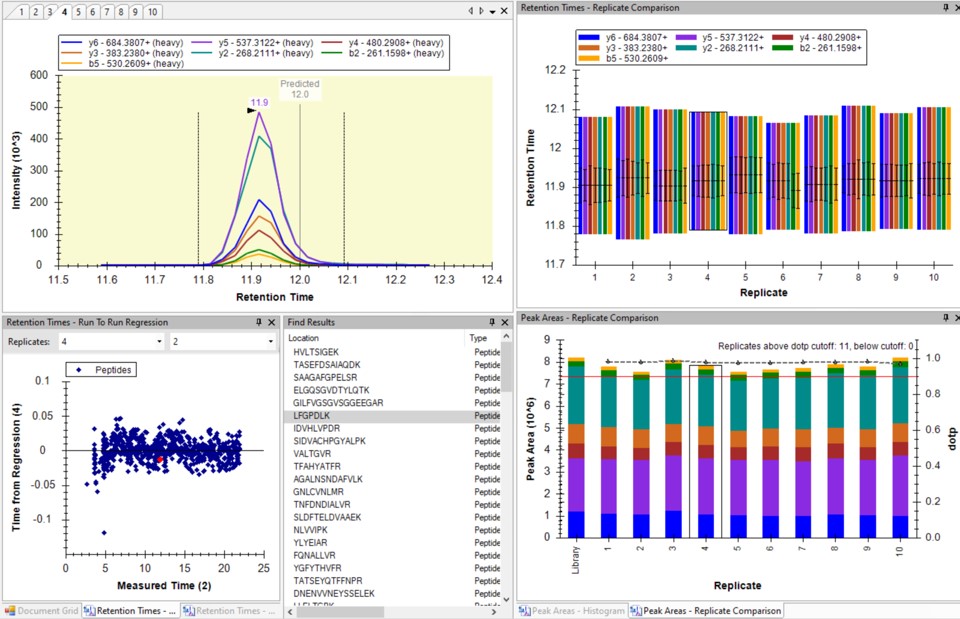
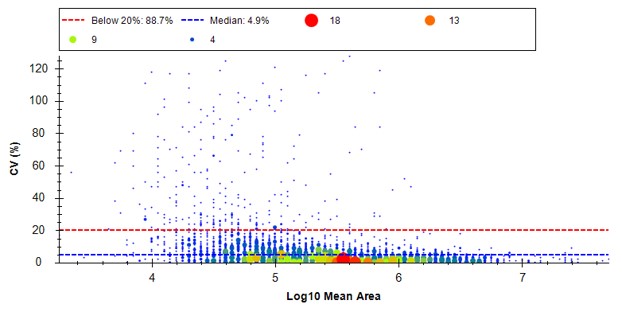
Select View / Peak Areas / CV Histogram. The CV histograms have ~94% of the targets with CV < 20%, with medians of 3.8 and 4.9%, which are excellent.

You can click on the histogram, which will open a Find Results window with some of the peptides that are close in CV to the value that was pressed. Double clicking any peptide sequence in the Find Results table will make that peptide active, whereupon one can check the peak shape, peak area, and retention time variations for the 8 replicates. Many/most peptides have results like LFGPDLK below.
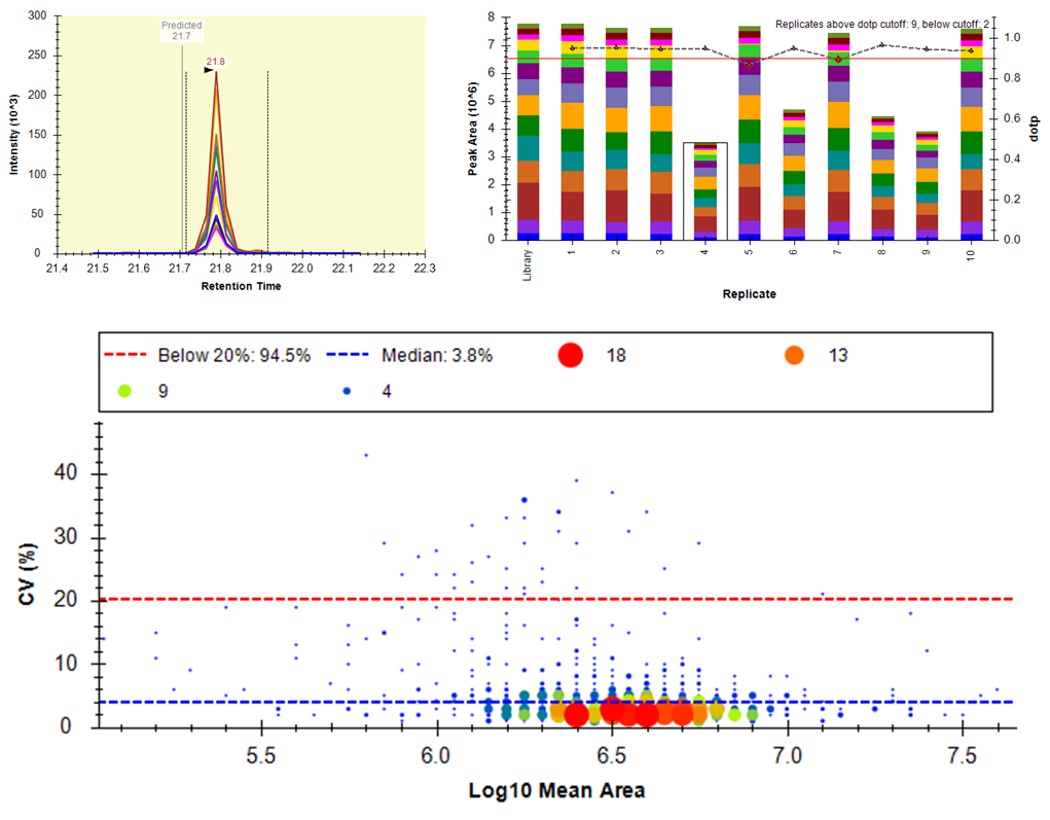
- Another interesting plot is the 2D CV Histogram, found under the View / Peak Areas menu. Here we see that there are 45 targets with CV > 20%. Clicking on the dots makes the peptide in question active. An example peptide SLADELALVDVLEDK is shown below, that has > 20% CV. It has a very skinny peak shape, eluting during the column wash portion of the run. Our analyses show that peptides very early and late in the assays have a high probability of have poor CV. One way to filter these peptides out is based on their narrow peak shape. PRM Conductor can set a minimum LC peak width bounds to do this.
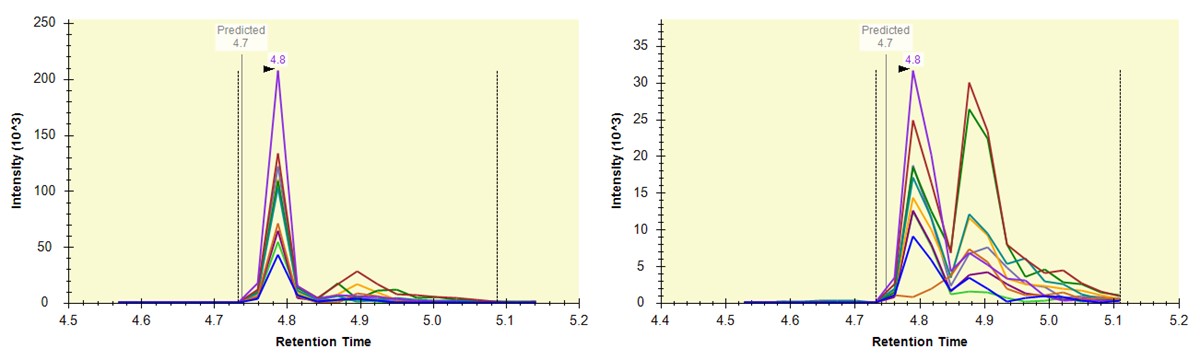
- Some peptides have bad CV's because they have interference an interference that varies from one replicate to another, like ANHEEVLAAGK below.
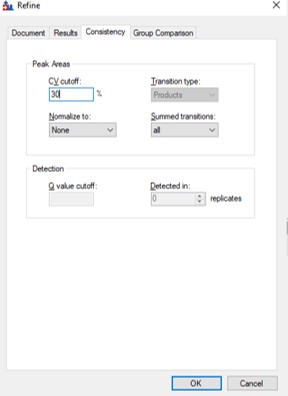
- One could simply filter all the peptides with a CV greater than a threshold from the document, using Refine / Advanced / Consistency, and set 20% in the CV cutoff box. Or one could try using MS3 acquisition in PRM Conductor, to save those peptides with poor CV's.
Analysis of Both Light and Heavy Peptides
-
Now we will add in the light precursors, that were measured but currently are not in the Skyline document. Save the document and then Save again with the names pq500_60spd_plasma_final_lightheavy_replicates.sky and pq500_100spd_plasma_final_lightheavy_replicates.sky.
-
Use Refine / Advanced, and select the Add box. The Remove label type combo box title changes to Add label type. Select light and press Okay to close the Refine dialog. Each peptide will now have its light precursor added. Use Edit / Manage Results, select all the replicates and press the Reimport button.
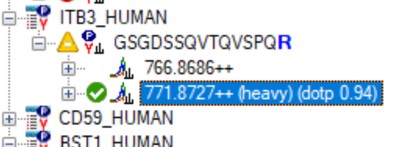
- Use View / Peak Areas / 2D CV Histogram, and set Normalized to Heavy. The PQ500 standards were designed to look for a set of proteins of interest, not all of which are expressed in normal plasma. For this reason many of the Light / Heavy area ratios are low, and have high coefficient of variance.

- Some peptides, like AGALNSNDAFVLK have significant endogenous peptide, and therefore have very good CV. Other peptides like EILVGDVGQTVDDPYATFVK have no observable endogenous peptide, and the area ratio therefore has a much higher CV. This general trend is observed in the CV histogram above.

-
This is the end of Step 4. We've demonstrated how to analyze replicate data for absolute quantitation with light and heavy peptides. A next step that some users will want to perform is a dilution curve. For absolute quantitation this takes two forms.
- Heavy Dilution: Constant endogenous (light) peptide amount, varying heavy spike-in concentration
- Light Dilution: Constant heavy spike-in concentration, varying endogenous sample.
-
The Light Dilution is a little easier to perform, because with the Settings / Peptide Settings / Modifications / Internal standard type is set to heavy, and thus Skyline uses the integration boundaries of the heavy peptides to integrate the light signals and determine whether the light/heavy ratios are sufficient for quantitation.
-
The Heavy Dilution is difficult, because eventually Skyline can't find the heavy peptide signal, and doesn't keep a constant integration boundary. Sometimes Skyline will jump over to the next biggest LC peak and ruin the dilution curve. We have sometimes used a script to set constant integration boundaries and solve this issue.
-
Calculating LOQs and LODs for large scale assays is still a little difficult, and we have used python scripts to do this. Skyline is also working on making improvements, and there will be updates in the future. We are submitting a paper soon that will have links to these scripts, for the intrepid that might be interested in exploring them.
Label Free - E. coli
E. coli Assay Development Overview
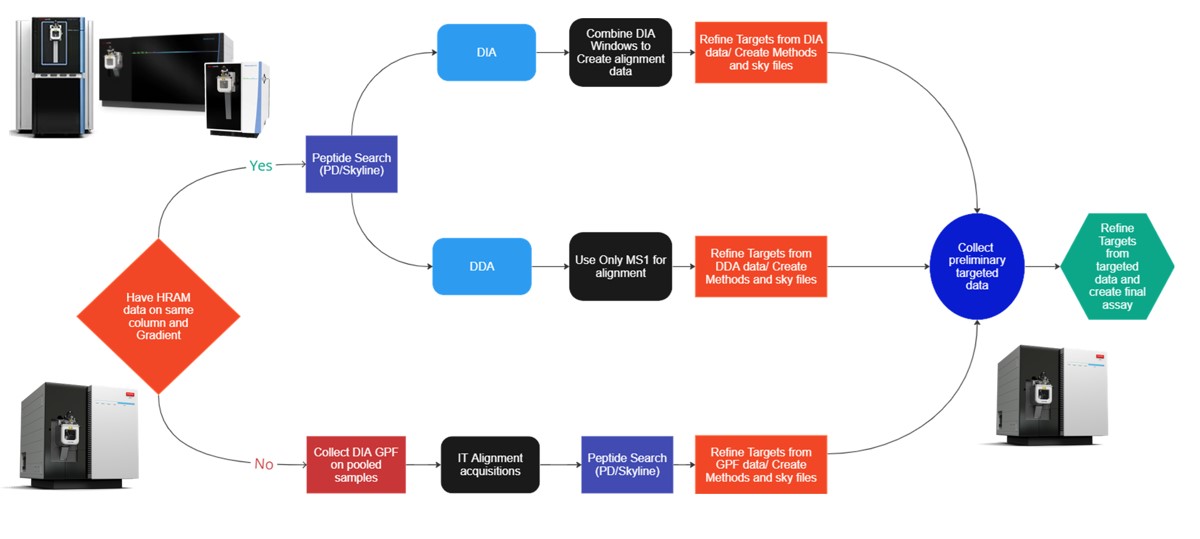
This tutorial will show you how to create a label-free targeted assay from discovery results. As shown in the figure below, there are two main routes to creating this kind of assay, to main difference being whether the discovery data comes from a high resolution accurate mass instrument like Astral, Exploris, or a Tribrid, or whether the discovery data comes from a Stellar. Once we have the discovery results in Skyline, the steps are largely the same. In this tutorial we'll look at an assay created from Stellar MS gas phase fractionation results.
Collect Gas Phase Fractionation Data on Pooled Samples
Overview of the Library Methods
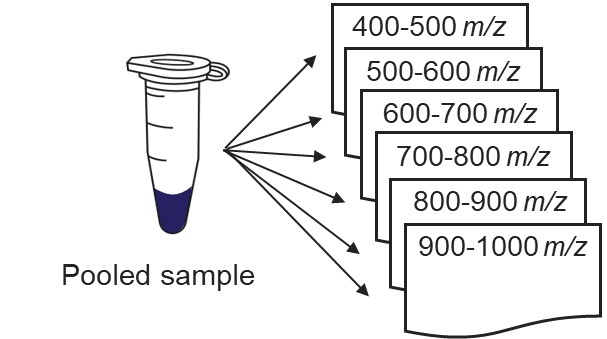
Our recommended technique for creating targeted assays starts with acquiring a set of narrow isolation window DIA data on a pooled or characteristic sample, creating what the MacCoss lab has termed a “chromatogram library”. This technique allows for a deep characterization of the detectable peptides in the sample, and the resulting data can be used to create a Skyline transition list of high quality targets. As outlined in in the figure below, generation of the library typically involves several (~6) LC injections, where each injection is focused on the analysis of a small region of precursor mass-to-charge. This technique has also been called “gas-phase fractionation” (GPF) to contrast with the traditional technique of off-line fraction collection and analysis. The GPF experiment should be performed with the same LC gradient as the final targeted assay, which allows us to use the library data for real-time retention time alignment during later tMS2 experiments.
Assuming that a pooled sample is available for analysis, the first step is to create a template instrument method file (.meth). Open the Instrument Setup program from Xcalibur, or the Standalone Method Editor (no LC drivers) if using a Workstation installation.
C:\Program Files\Thermo Scientific\Instruments\TNG\Thorium\1.0\System\Programs\TNGMethodEditor.exe
Use the method editor to open the file at Step 1. DIA GPF/60SPD_DIA_1Th_GPF.meth. This method includes 3 experiments:
- An Adaptive DIA experiment for retention time alignment that has a wide isolation window and ultrafast scan rate. We use Window Optimization Off so that later these windows will not be imported into Skyline, which can be accomplished because both the analytical DIA experiment (experiment #3) and later targeted methods will not use integer precursor m/z values, and Skyline will use a 0.0001 Th tolerance.
- An MS experiment, used for diagnostic purposes. Some DIA processing algorithms may also use the MS data for peptide searching, but currently nominal mass MS data don’t appear to be useful for DIA. Note that MS data can be used for data normalization in Skyline, even when MS data are not being imported.
- An analytical DIA experiment for peptide searching that use a narrow (~1 Th) isolation window. Window Optimization is set to “On” so that isolation window edges are in “forbidden” m/z zones and we don’t have to use any window overlap.
Assuming that the user has configured their LC driver with the Instrument Configuration tool, the LC driver options would appear as a tab in the pane on the left-hand side of the method, where the user has to design the length and type of gradient program to be used. Of course, the choice of gradient length is of great importance as it determines the number of compounds that can be analyzed and the experimental throughput. A general rule of thumb is that Stellar can analyze on the order of 5000 peptides in an hour gradient, where this number depends on how the peptide retention times are distributed and other factors and settings that will be explored later in the tutorial. The user will need to make sure that the MS part of the experiment has the same Method and Experiment Durations as the LC gradient.
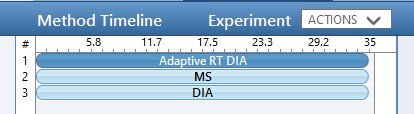
While we are making methods, we should create a tMS2 method template with the same LC parameters as the library method file. One can start with Step 1. DIA GPF/60SPD_tMS2_Template.meth. This method has the same first two experiments as the GPF template, and the 3rd experiment been replaced with a tMSn experiment. Our software tool will later fill in the targeted table information in a new version of this file, as well as update the LC peak width and cycle time parameters and embed the reference data for the Adaptive RT.
One thing to note about the targeted template is that the Dynamic Time Scheduling is set to Adaptive RT. When we save the method, a message tells us that the file is invalid because no reference file has been specified. This is okay, because PRM Conductor will fill in this information for us later.

Creating the GPF Methods
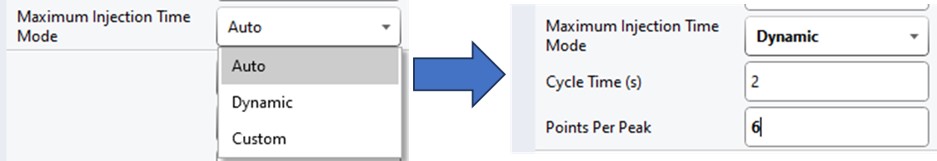
Once the GPF method template file is created and saved, the user should select that file with the GPF Creator tool. The tool will create a set of new methods based on this template, one for each precursor mass-to-charge region. The number of method files to be created depends on the settings in the user interface. The default precursor range for the tool is 400 to 1000 Th, as most tryptic peptides have a mass-to-charge ratio in this range. We recommend, for Stellar, to use an isolation width of 1 Th, which allows the DIA data to be searched even with traditional peptide search tools like SEQUEST and allows for a good determination of which transitions will be interference free in the tMS2 experiment. However some users have used 2 Th windows, in order to user slower scan rates and/or more injection time per scan without having to acquire 2x as many GP fractions. If the experiment Maximum Injection Time Mode is set to Auto, then the maximum injection time is the largest value that does not slow down acquisition, which depends on the Scan Rate. For this simple tool, we assume a scan range of 200-1500 Th. The largest amount of injection time and the slowest acquisition rate is afforded by the 33 kDa/s scan rate, and the smallest amount of injection time and the fastest scan rate is afforded by the 200 kDa/s scan rate.
For most applications, we prefer to use 67 or 125 kDa/s and set the Maximum Injection Time Mode to Dynamic as in the figure below, which lets the maximum injection time scale with the time available in the cycle.
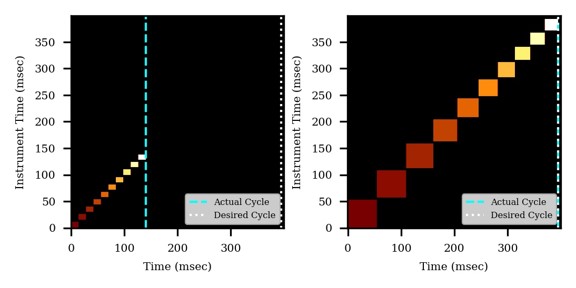
If the Cycle Time is set to 2 seconds and the cycles take less than 2 seconds, then the remaining time in the cycle is distributed to the acquisitions inversely proportionally to the intensity of the precursors, ex. less intense precursors get more injection time. In this figure, the left panel shows a hypothetical situation at the start of a cycle. We calculate the minimum amount of time that each of the acquisitions needs, and sum this to get the blue, vertical, dashed line. The remaining time in the cycle is the white, vertical, dotted line. Each acquisition is colored, with the most intense precursors colored white, based on previous data. In the right panel, the amount of time the acquisitions actually take is displayed, where the less intense precursors were given more injection time.
The expected peak width in GPF Creator is the LC peak width at the base in seconds. This value can be determined by checking a few peaks in a quality control (QC) experiment, for example an injection of the Pierce Retention Time Calibration standard. The LC peak width combined with the desired number of sampling points across the peak width, the isolation width, and the Scan Rate determines the number of LC injections needed to create the library.
Once the parameters are set, the user selects the Create Method Files button, which will create the instrument method files needed for the library. The user can create a sequence in Xcalibur and run these methods along with any blanks and QC’s that they want. We have collected data for a mix of 200 ng/ul E. coli with 200 ng/ul HeLa, and placed the resulting raw files in Step 1. DIA GPF\Raw.
Search Library against Organism Database
The next step is to search the data to find candidate peptide targets. Ion trap DIA data with 1 Th isolation windows can be searched for peptides using Proteome Discoverer and the SEQUEST or CHIMERYS search engines. We have included example templates for Proteome Discoverer with this tutorial. SEQUEST in Proteome Discoverer was built to handle data dependent acquisition experiments, and so a few parameters are set to unexpected values. The Precursor Mass Tolerance is set to 0.75 Da, which means that each spectrum is queried against the peptides in a window that is +/- 0.75 * charge_state. For the spectrum selector node, we set Unrecognized Charge Replacements to 2; 3, so that each spectrum is searched as both charge state 2 and 3, because Stellar DIA acquisitions are marked with Charge 0, which is Unrecognized for PD. This practice, plus the wide precursor mass tolerance, means that each spectrum in the chromatogram library takes a long time to search; on the order of an hour for a 30 minute gradient method using a standard instrument PC, ex 6 hours to search a GPF experiment. CHIMERYS searching is often much faster for this kind of experiment.
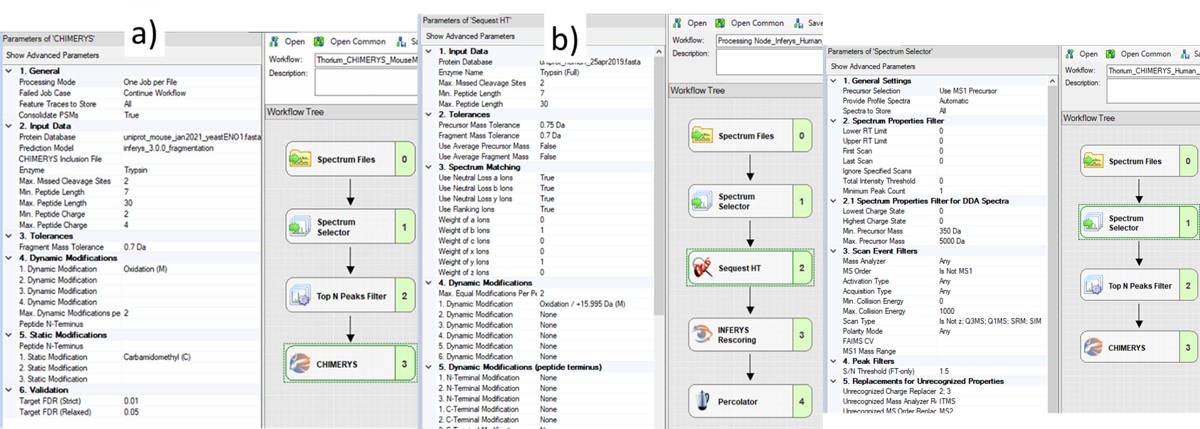
Also, in the spectrum selector node we set the Scan Type to “Is Not Z” (only Full), so that the large isolation width Adaptive RT acquisitions are filtered, which have a Z in the scan header.
A nice, new feature in the Proteome Discoverer 3.1 SP1 is the addition of the Consolidate PSMs option in the Chimerys 1. General section. Setting this parameter to True tells Chimerys to keep on the best PSM per peptide/charge precursor. This consolidation has the effect of speeding up the Consensus workflow significantly, as well as reducing output file size and the amount of time needed for Skyline to import the results.
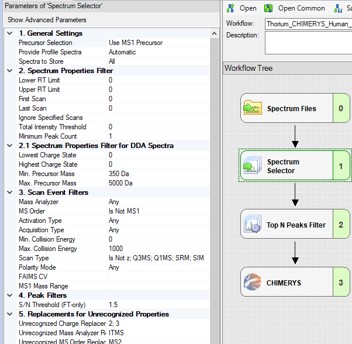
To process the files in Proteome Discoverer, one can first create a study (a), set the study name and directory, and the processing and consensus workflows (b). The workflows can also be set or modified once the study has been created. When the study has been created, one uses Add Files to choose the library raw files, and drags them to the Files for Analysis region, and then presses the Run button to start the analysis (c)
Manual Import Search Results into Skyline
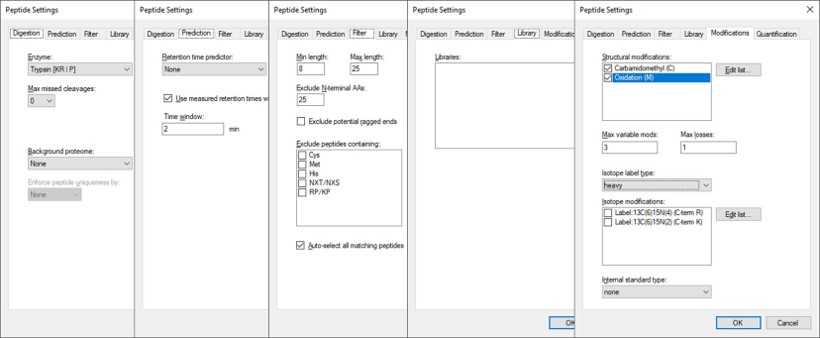
After Proteome Discoverer finishes processing the data, a file is produced with the .pdResult extension, and another one with the .pdResultDetails extension. Both files must be in the same folder for Skyline to import the search results and to start refining a First-Draft targeted method. We will first show you how to import the results manually, and then we will present a simple tool to perform the import with the settings that we recommend for ion trap analysis. To import the PD results manually, create a new Skyline document, and save it as ImportedGPFResults\gpf_results_manual.sky. Open Settings \ Peptide Settings, and set the parameters as in the figure below. The main points are to ensure that no Library is selected, and the Modifications do not specify any isotope modifications. Skyline will check for Structural modifications in the imported search results and ask you about any that are not already specified. Press Okay on the Peptide Settings dialog box.
Open the Settings \ Transition Settings and use settings like below. Set Precursor charges to 2, 3, 4, Ion charges to 1,2, and Ion types to y,b. Don’t specify ‘p’ in Ion types, because PRM Conductor currently takes this as a sign that DDA is being used and loses some functionality. Set Product ion selection from ion 2 to last ion. For Ion match tolerance use 0.5 m/z, with a maximum of 15 product ions, filtered from product ions. Set the Min m/z to 200, and Max m/z to 1500 (whatever was used for acquiring the GPF data), with Method match tolerance of 0.0001. Set MS1 filtering to None, and MS/MS filtering to Acquisition method DIA, Product mass analyzer QIT with Resolution 0.5. For the Isolation scheme, if a suitable scheme is not yet created, select Add. Here we set 0.35 for the Retention time filtering, but a wider window may be more acceptable for longer gradients.
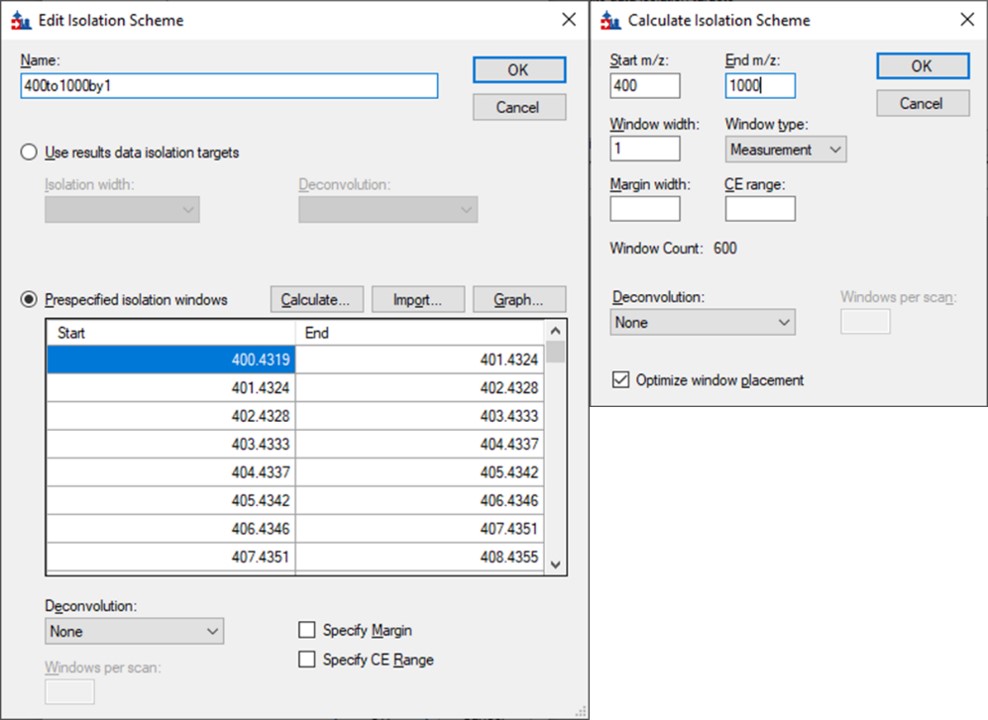
As below, once Add is selected for Isolation Scheme, give it a suitably descriptive name, then press Calculate. Set the Start and End m/z to the limits and isolation width used in the GPF experiment, and click the Optimize window placement button, assuming you used that option in the GPF experiment. Press Ok twice to close the Isolation Scheme Editor, and once more to close the Transition Settings.
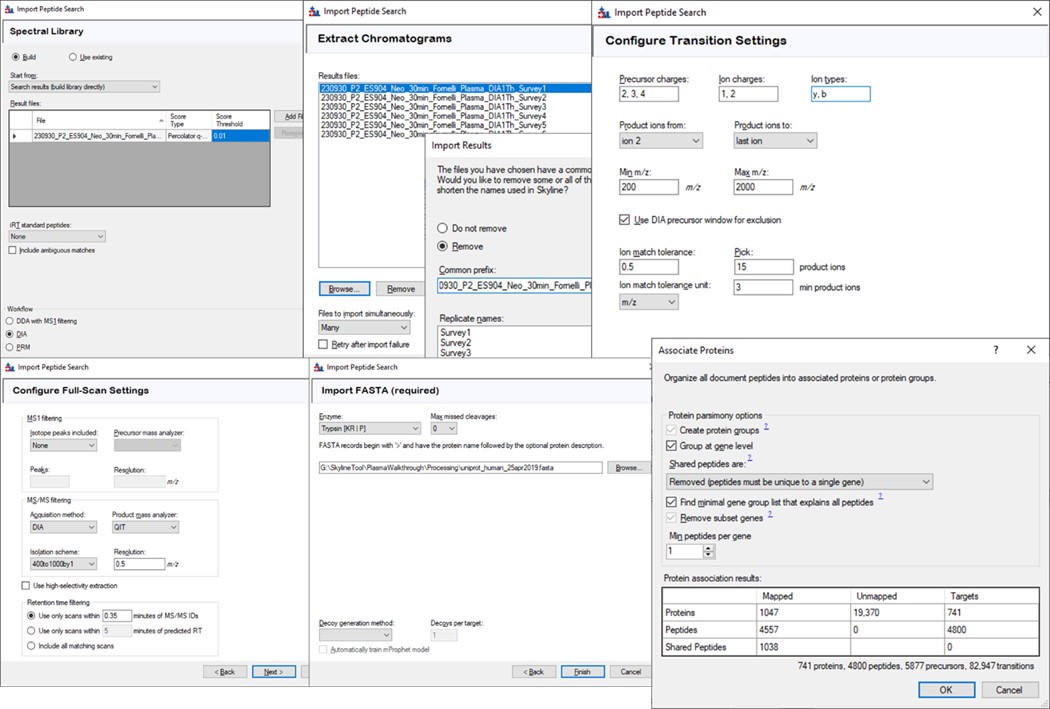
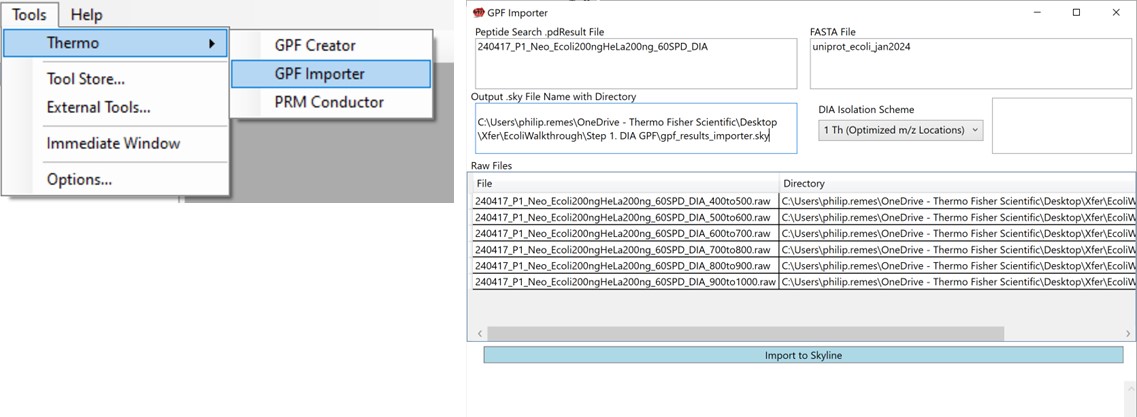
Save your Skyline document at this point, and select File / Import / Peptide Search. Use the Add Files button to navigate to and select the Step 1. DIA GPF/Processing/240417_P1_Neo_Ecoli200ngHeLa200ng_60SPD_DIA.pdResult file.
As in Figure below, select Score Threshold 0.01, Workflow DIA and press Next. Skyline will build a .blib file from the .pdResult, which will take several minutes. If you get an error during this step, ensure that the .pdResultDetails file is also present in the same folder as the .pdResult file, for PD >= 3.1, and ensure that you are using an updated version of Skyline (preferably the Daily version). When this step finishes, Skyline presents a .raw file chooser dialog, select Browse and select the files in Raw / GPF. Press Next, then you can remove or not remove portions of the file names to shorten them, and press Okay. The next Add Modifications tab should be empty if the Peptide Settings / Modifications were already specified, so press Next again. In the Configure Transition Settings, Skyline will have overwritten several of your Transition settings, so remove the ‘p’ from Ion types, and set Ion match tolerance to 0.5, and Max product ions 15, Min 3 and press Next. In the Configure Full Scan Settings, set the MS1 Isotope Peaks to None, and press Next. In the Import FASTA tool, Browse and select the .fasta file in the Processing folder. You can set the Decoy generation to None, especially if you used PSM Consolidation to reduce to a single PSM per peptide, then press Finish. Skyline will start associating peptides to proteins now, displaying a progress bar, and eventually shows an Associate Proteins tool. Here the most strict setting would be to create protein groups and remove any peptides that are not unique to a protein, but alternatively one might choose to Assign to the first gene, for example. Press Okay, and Skyline will import the results from the .raw file, which may take a few minutes. Note that one could have skipped the results importing and pressed cancel at any time after the library was created. The library would be available to browse with View / Spectral Libraries, and the peptides could be added to the document with Add All. Occasionally this process can be used if there were any errors or issues with modifications. Additionally one could use this strategy to load the .raw files as a single multi-injection replicate, the way that the GPF Importer will do, which makes viewing the results much more convenient as we’ll see below. When the results are finished loading, save the gpf_results_manual.sky file.
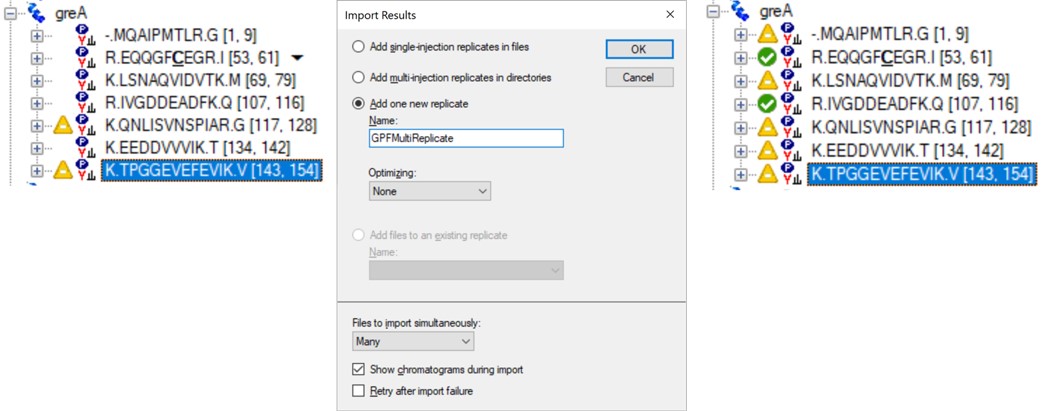
Click on the second gene labeled greA to expand it. Notice that with the 600to700 file selected, only two peptides are visible. With the scheme we have used, each .raw file was loaded on its own, instead of as part of a whole. To see the difference, use Edit / Manage Results (or Ctrl+R) to open the Manage Results tool, press Remove All and Okay. Now use File / Import / Results. When asked if you want to use decoys say No. In the Import Results tool, select Add one new replicate and use the name GPFMultiReplicate, and press Okay. Choose the six .raw files in the Raw / GPF folder again, and they will be immediately imported if you had finished the importing with the Wizard. The greA gene that is expanded now will have all its peptides completely filled in, like below. Save the gpf_results_manual.sky document.
GPF Import Search Results into Skyline
We’ll now look at how the GPF Importer does the same thing as above, but with fewer steps. To use the GPF Importer tool use Tools / Thermo / GPF Importer. Double click the Peptide Search and FASTA file boxes to select the .pdResult file and FASTA file respectively. In the Output file Path, Copy/Paste in the output directory you want, and type an appropriate name for the new Skyline file. Double-click the Raw File pane and select the associated GPF .raw files and press the Import to Skyline button. The log window shows the progress of the import, which takes several minutes. When it finishes, the result will be the same as in the figure below when the multi-injection replicate was imported.
This is the end of the first step. We have Skyline files with peptide search results in them. In the next steps we'll refine the results on our way to making a final assay.
Validation with Subset Replicates
- Refining the Search Results
Refining the Search Results
Use PRM Conductor Skyline plugin to refine the transitions and precursors
Launching PRM Conductor
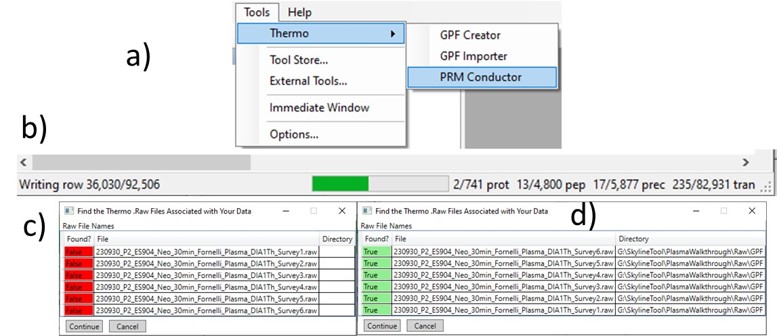
Once the results are imported into Skyline, the next step is to create a First-Draft version of the targeted assay using the PRM Conductor Skyline plugin. If you followed the steps in the tutorial, the Step 1. DIA GPF / gpf_results_importer.sky file should have opened after the import was finished, or you have the gpf_results_manual.sky version made. From one of these files, launch the PRM Conductor program (a), and Skyline’s bottom bar will report the progress of creating a Skyline custom report file that gets loaded by PRM Conductor (Figure b). Note that PRM Conductor depends on .raw files for some analysis, so it will look in obvious locations for the data files referenced in the Skyline file. If the raw files were not found, it will ask you to find the missing files. Once you double-click and use the file chooser to select them, the dialog will show a green “True” and you can continue. Even if the Skyline file used .mzml or another raw file format, just place the associated .raw files in a nearby folder and you can use the PRM Conductor.
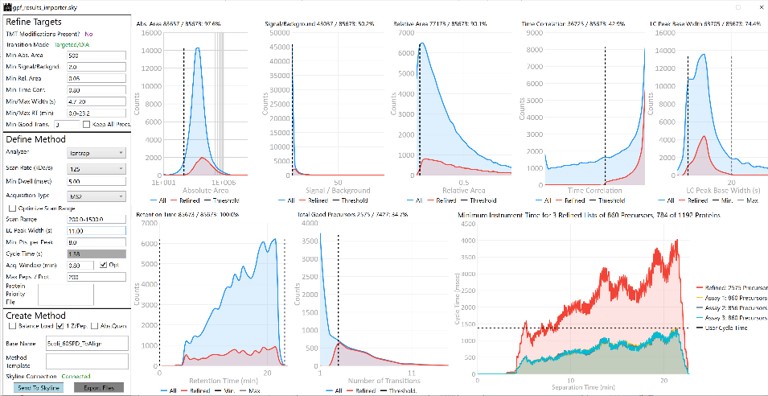
Once loaded, the user interface in the figure below will be shown. There are 3 main parts to the user interface: a set of parameters on the left, a set of transition metric plots on the top right, and a set of precursor metric plots on the bottom right. We start with the Refine Targets parameters and the associated plots. Each of the text boxes has a corresponding graph on the right. The current value in the text box is a threshold value, and is displayed with the graph as a dashed, vertical line. The title of the plot reports how many of the transitions were filtered based on the current threshold. The blue distribution in the plot is for all the transitions in the report, while the red one is after all filters have been applied. Changing the parameter in the text box and pressing Enter will update the calculations in the plots on the right.
Refine Targets Section
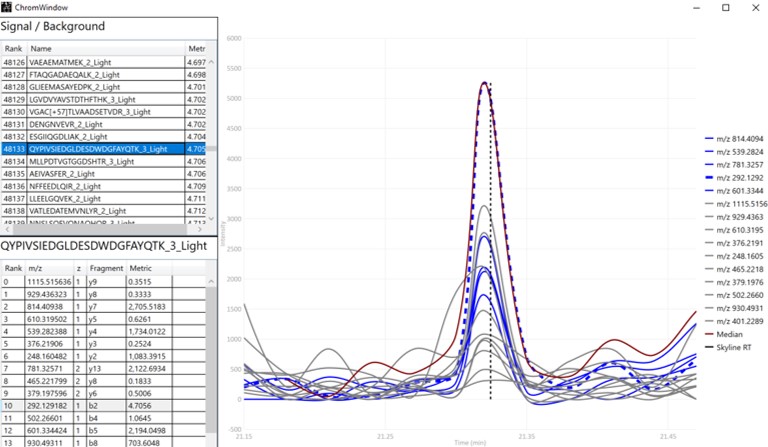
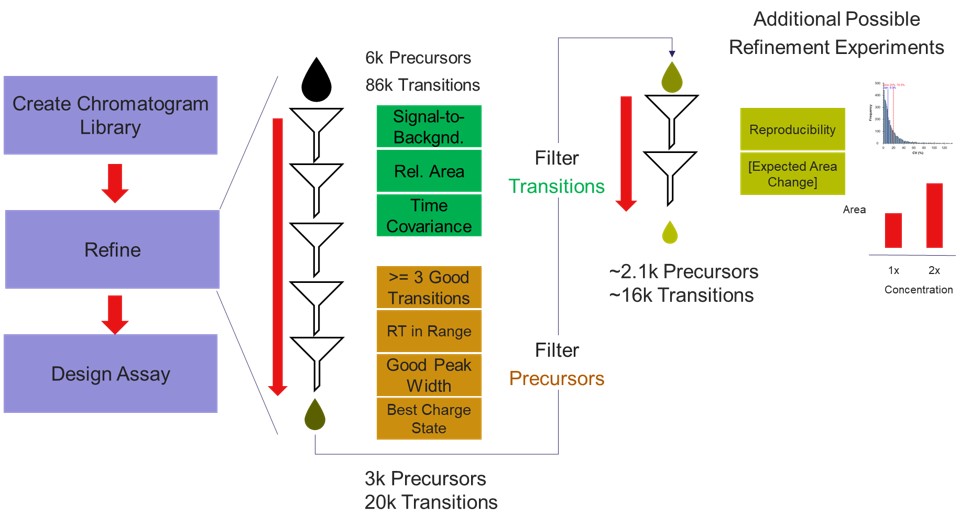
The first plot on the top is Absolute Area distribution. These are the Area values determined by Skyline for each transition. Note that these values are analyzer dependent, so a threshold used for Orbitrap discovery data may not be suitable for the Ion trap discovery data. The second plot is the Signal/Background distribution. These are the values of Area and Background as determined by Skyline for each transition. A traditional threshold for S/B is 3, however the user may wish to adjust to a different value. Highlighting the blue distribution and left-clicking will bring up a new window that gives the user an idea of what a particular S/B looks like in the Skyline transition data. For example, the figure below is a view of the plot after clicking the Signal / Background plot around 4. In the top left, all the transitions are in a grid, sorted by Signal / Background. On the bottom left is a grid showing all the transitions for the peptide corresponding to the selected transition on the top left grid. The graph on the right shows all the transitions for this peptide. The selected transition is highlighted in dashed lines. All transitions that currently meet the thresholds are colored blue, while the other transitions are colored grey. The red transition is the median transition trace.
The third plot on the top is the Relative Area distributions, that is, the area normalized to the largest transition for each precursor. The fourth plot is for the time correlation of each transition to the median transition for each precursor. The fifth plot on the top is for width of the transitions at the base, where a general rule is that outliers from this distribution may be poor quantitative markers, either because they are too narrow to characterize with the same acquisition period as wider peaks or are wide and potentially not reproducible. On the bottom row, the first plot is the retention time distributions. Precursors that are at the very beginning and very end may have the highest variability and could potentially be filtered out. The final transition filtering parameter is the Min Good Transitions text box. Many researchers prefer to set this value in the range of 3-5, to increase the confidence that a set of transitions is a unique signature for a peptide of interest. However, you could also click the Keep All Precs checkbox if you wanted to keep all the precursors, along with their best Min Good Transitions, which would potentially include some “bad” transitions. This is useful for cases with heavy standards where you don’t want to remove any of them. This is used in the Absolute Quantitation - PQ500 walkthrough.
Define Method Section
The next set of parameters on the left, in the Define Method box, control the acquisition parameters to be used in the targeted method. These parameters affect the speed of acquisition and the number of targets that can be scheduled in an analysis. The first parameter in the Define Method box is Analyzer. In this tutorial we are focused on Ion Trap analysis, but it is possible to make assays for other Analyzers. Selecting an Analyzer makes a specific set of parameters visible, which can change the characteristics of the analysis.
When any of these parameters are updated, the Scheduling graph on the bottom right will update. This graph shows the distribution of retention times for all Refined precursors in the red trace and the user’s chosen Cycle Time with the horizontal, dashed, black line. As we will see later, the user can choose to create a single assay that schedules as many precursors as will fit beneath that Cycle Time or may choose to create as many assays as necessary to acquire data for all refined precursors. The figure below shows views of the Scheduling graph in the single assay or “Load Balancing” mode, which we will talk more about below.
For peptide analysis, there is not much practical utility for the additional resolution afforded by the 66 kDa/s or 33 kDa/s over the “unit mass resolution” of the 125 kDa/s scan rate. Because for targeted methods we typically use the Dynamic Maximum Injection Time Mode in the instrument method file, the instrument allocates any additional time in the cycle to the targets according to their intensity. Therefore, the slower scan rates historically were useful mostly to guarantee a minimum amount of injection time per target, above the ~13 ms afforded by 125 kDa/s with the 200-1500 Th Scan Range, which can be useful for low concentration samples. With PRM Conductor, one can also set a minimum injection (or dwell time for QQQ users), which can accomplish the same thing. For peptides, we typically use the fixed 200-1500 Th scan range, because it guarantees this amount of injection time, gives a reasonable acquisition speed (~65 Hz), and the fragment ions in regions above and below this range are not essential to characterize most tryptic peptides.
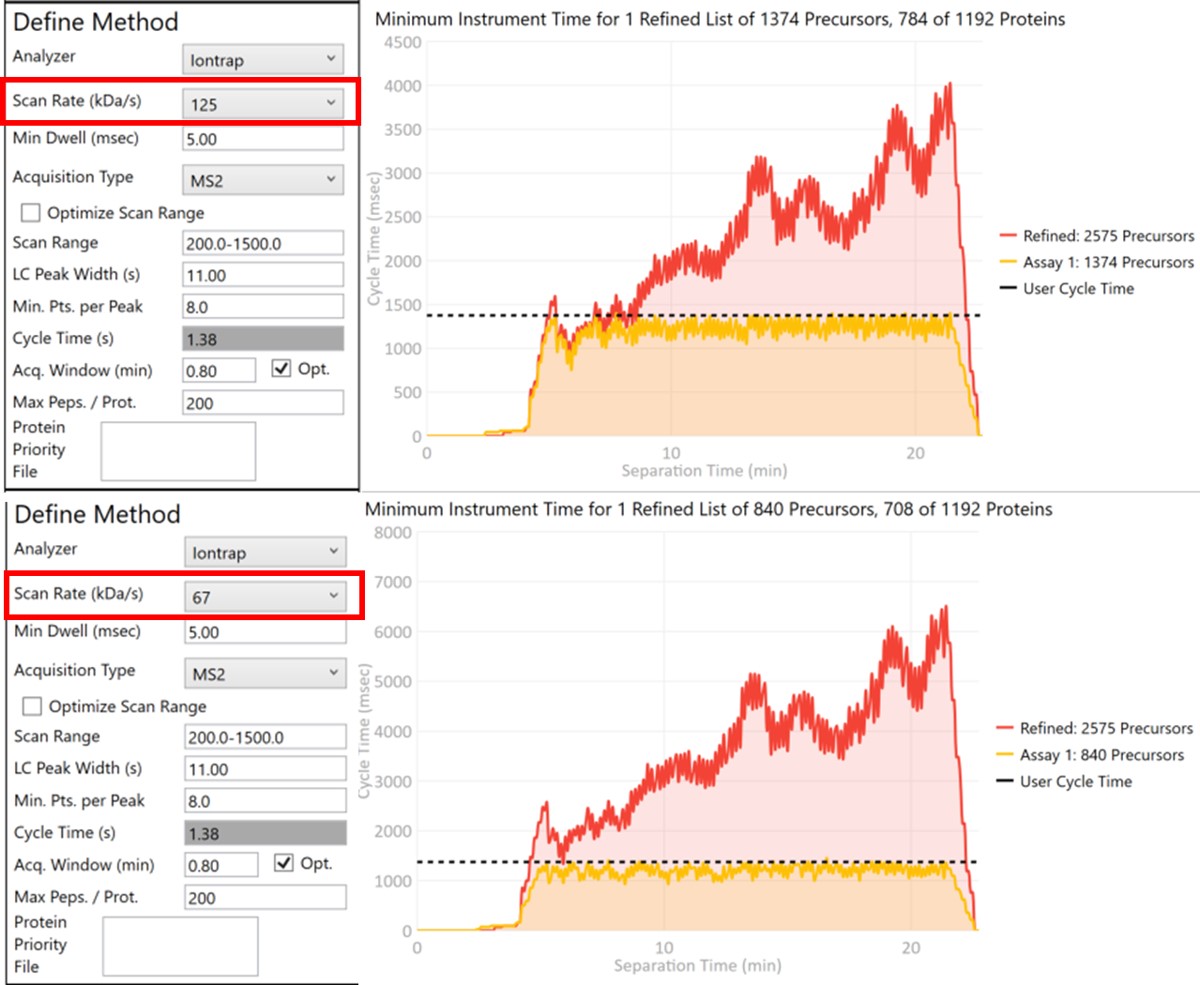
Let’s see what effect the Scan Rate has on the experiment. Changing the Scan Rate updates the estimation of how fast the instrument can acquire data. This is reflected in the Minimum Instrument Time plots, where in the top view, the Scan Rate is set to 125 kDa/s. An assay that would acquire data for all 2575 Refined Precursors would take 4 sec at the peak (at ~21 min), and 1374 precursors are able to be scheduled with the user-defined Cycle Time of 1.38 seconds. When the Scan Rate is set to 66 kDa/s (bottom part of the figure) the 2575 Refined Precursors would take 6.5 seconds of instrument time, and only 840 precursors can be scheduled with a Cycle Time of 1.38 seconds.
The user can change any of the parameters in this pane and visualize the effect on the assay. Of particular importance is the Acquisition Window. Stellar instruments are enabled with a new algorithm for real-time chromatogram alignment called Adaptive RT, which allows the instrument to adjust when targets are acquired to account for elution time drift. We find that an Acquisition Window setting of 0.75-1.00 minute usually results in good data, at least for the gradients we have typically used, in the range of 30 to 60 minutes. As LC peak width decreases for shorter gradients, we have successfully used narrower Acquisition Windows of 0.5 or even 0.3 minutes. Traditional tMS2 experiments, without real-time alignment, can sometimes require Acquisition Windows in the range of 3-5+ minutes. Adjusting this parameter allows to visualize the gain in number of targets afforded by narrower Acquisition Windows. For example, a 1 minute versus a 5 minute Acquisition Window allows to analyze more than 3x more targets (1172 versus 305). An additional feature related to Acquisition Window is the Opt check box. We have observed that the most variable part of many separations is at the beginning, which poses additional challenges for a real-time alignment algorithm because few if any compounds are eluting at the beginning. The Opt checkbox expands the Acquisition Windows somewhat, while still respecting the user’s Cycle Time requirement. The targets that are most improved by this algorithm are the early and late eluting targets; those where expanding their acquisition windows has little effect on the available injection time, but where alignment can be the trickiest.
The last two parameters, which will be explored more below, are the Max Peptides per Protein, which helps to focus the analysis on just the highest quality peptides from each protein, and the Protein Priority File, which allows to create exceptions for particular proteins using their Skyline protein names.
Create First-Draft Targeted methods and Skyline analysis file
Exporting Multiple Methods for All Refined Precursors
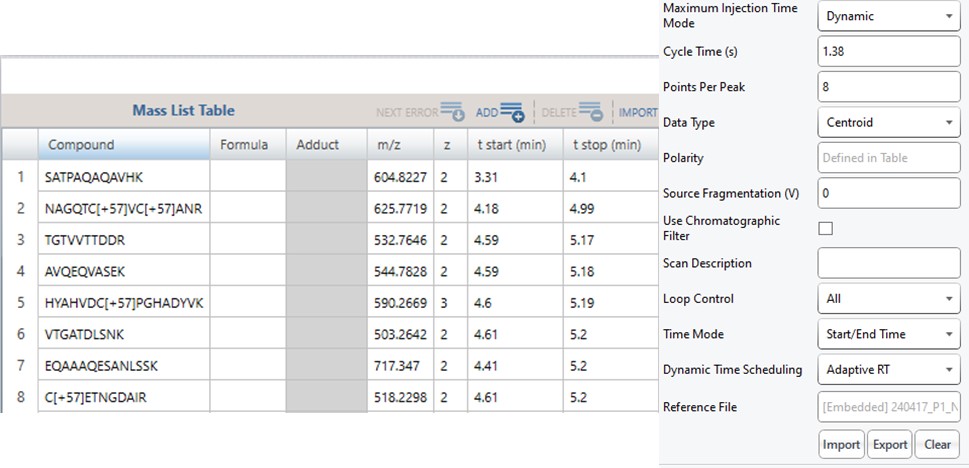
Now we will create multiple tMS2 methods for all the precursors that passed the filters and check to see which ones are stable and reproducible quantitative targets. To do this, uncheck the Balance Load button, which changes the Minimum Instrument Time plot so that all the Refined precursors are split into 3 assays, each of which require less acquisition time than the 1.38 second Cycle Time. Enter a base name for the file that will be created, otherwise the name “assay” will be used. Double-click the Method Template box and find the Step 2. Validation with Subsets/60SPD_PQ500PRTC_AlignTemplate.meth file. Then click the Export Files button.
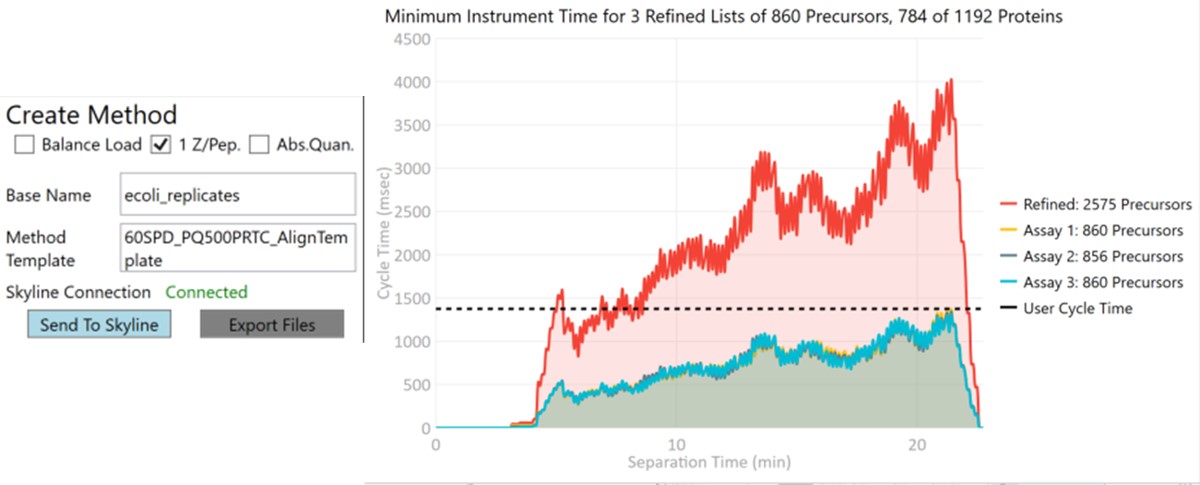
Some processing of the raw files will take place, as denoted by a progress bar, while the data files are aligned in time and a Adaptive RT reference file with the .rtbin extension is created. This processing only happens once, so if PRM Conductor is ever launched again for these raw files it won’t take as long to Export. The processing can take several minutes, depending on how many files are present and how long the gradient was. Method files will be created for each of the 3 assays in the same folder as the tempate method. A Skyline file will be created that is configured for tMS2 analysis with settings appropriate for the selected Analyzer. As a backup for the method and Skyline files, isolation and transition lists lists will be saved that are suitable for importing to the method editor or Skyline. The figure below is a view of the Step 1 folder showing some of the created files, and also the Step 2 folder showing the exported method files.
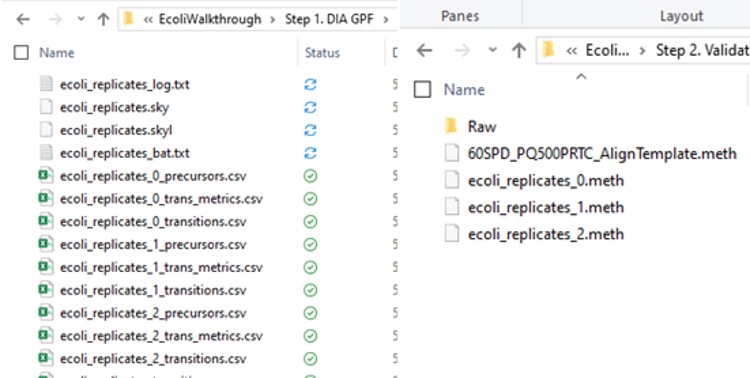
Note that because the 60SPD_PQ500PRTC_AlignTemplate.meth file had an Adaptive RT experiment and the tMSn method had Dynamic Time Scheduling set to Adaptive RT, the methods have embedded a .rtbin file to use for the real-time alignment. Notice too that the user’s Cycle Time and Points Per Peak were included, as well as the relevant precursors with their m/z, z, and scheduled acquisition times.
When exporting method files, a feature that can be useful is the Protein Priority File. There are two reasons to use this feature:
- when exporting multiple replicates, the peptides from any proteins on this list will be added to each of the replicates. The purpose of this is to support Skyline’s iRT feature, which requires the same iRT peptides to be present in each replicate. We explore this more in the walkthrough on absolute quantitation.
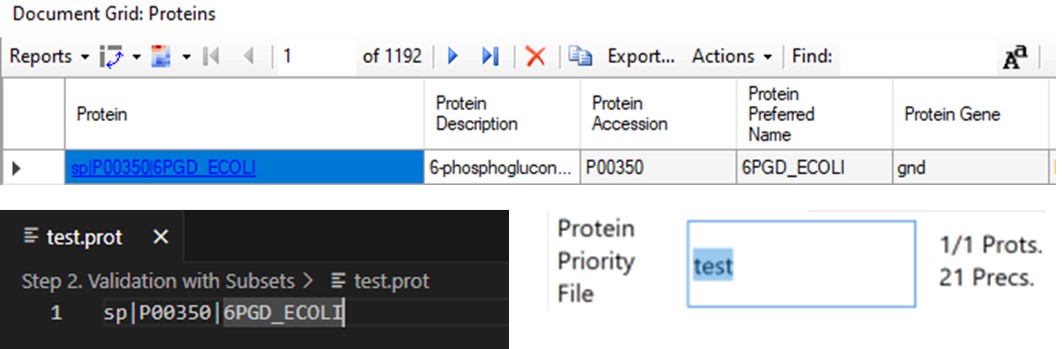
- When exporting a single replicate (balance load mode), any peptides from proteins on this list are added to the final list, assuming they passed the transition and precursor filters. To use the Protein Priority File feature, just create a text file with the .prot extension, and add any prioritized proteins to it, one per line, using exactly the format that Skyline uses for the Protein Name. For example, one could add sp|P00350|6PGD_ECOLI, as seen in the figure below in the Protein document grid, but not the Gene name displayed in the Skyline tree. When the .prot file is selected, the UI updates to display how many proteins and peptides are being prioritized. For this walkthrough we didn’t use the Protein Priority File feature, however.
Acquire First-Draft Targeted data on Stellar MS
If the user desired, they could rely on just the results from the library to create a targeted assay. However, because a targeted assay may be used for many samples, we have found it worth the extra effort to further validate the set of precursors for reproducibility by performing several injections with each of the first-draft assays and filtering on a minimum coefficient of variation (CV) on the LC Peak Areas. In some more advanced scenarios where the assay is part of a multi-proteome mixture, one could perform injections for at least two concentrations of the proteome of interest, to ensure that the selected peptides change area with concentration appropriately, and thus belong to that proteome. In this tutorial we will demonstrate filtering performed on the LC peak area CV.
Import First-Draft results to Skyline
We performed 2 injections for the 3 first-draft assays. The program was in a slightly different state at the time, so the data collected don’t perfectly match the Skyline file created in the previous step, but they are very close. Normally we could just use the ecoli_replicates.sky file created in the last step for importing the results, but because of the different program state, we'll instead do a Save As on the Step 1. DIA GPF/gpf_results_importer.sky and save a new file, Step 2. Validation with Subsets/ecoli_subset_replicates.sky.
- Change the Transition Settings / Full Scan / Acquisition method to PRM with QIT analysis, and set the Retention Time filtering to a value that will probably cover the retention time shifts in the coming weeks that this assay will be run, +/- 2 minutes.
- Use Edit / Manage Results and remove the Chrom_Lib_Replicate.
- Use File / Import / Results and select Add multi-injection replicates in directories, and then choose the Step 2. Validation with Subsets\Raw folder in the dialog that pops up. Press Okay and we can also Not remove the common 'Replicate' in the folder names. Loading the data as multi-injection replicates, them nicer to view in Skyline, because each of the multiple raw files involved in a replicate is merged into a single result, and peptides won’t show up as “missing” if the wrong result file is selected.
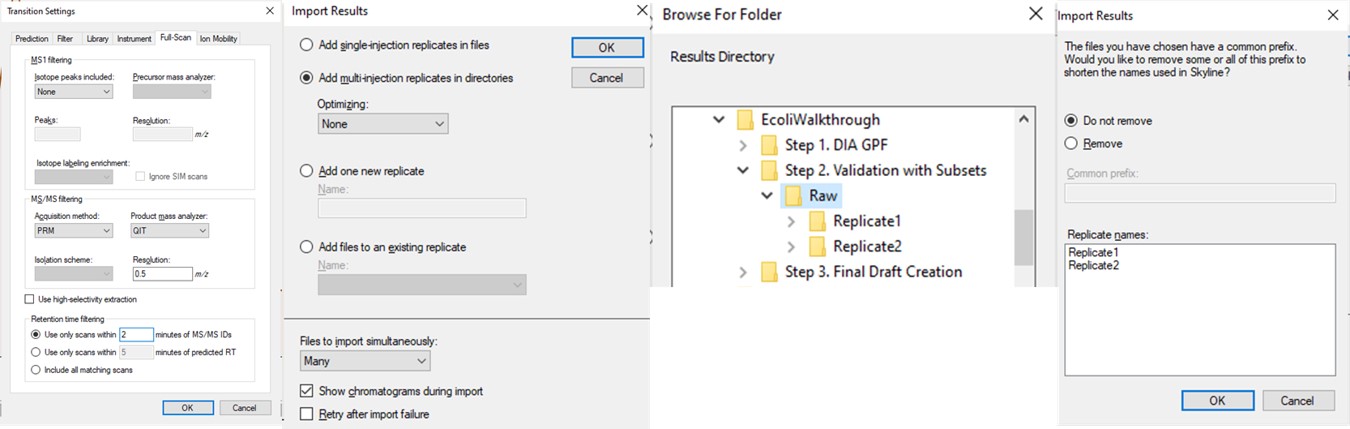
- Now though because we started from the gpf_results_importer.sky that has all the peptides, we should Refine / Remove missing results, and then Refine / Remove empty proteins.
- Arrange the graphs like in the figure below.
- View / Retention Times / Replicate Comparison
- View / Retention Times / Regression / Run-to-Run
- View / Peak Areas / Replicate Comparison
- View / Peak Areas / CV Histogram
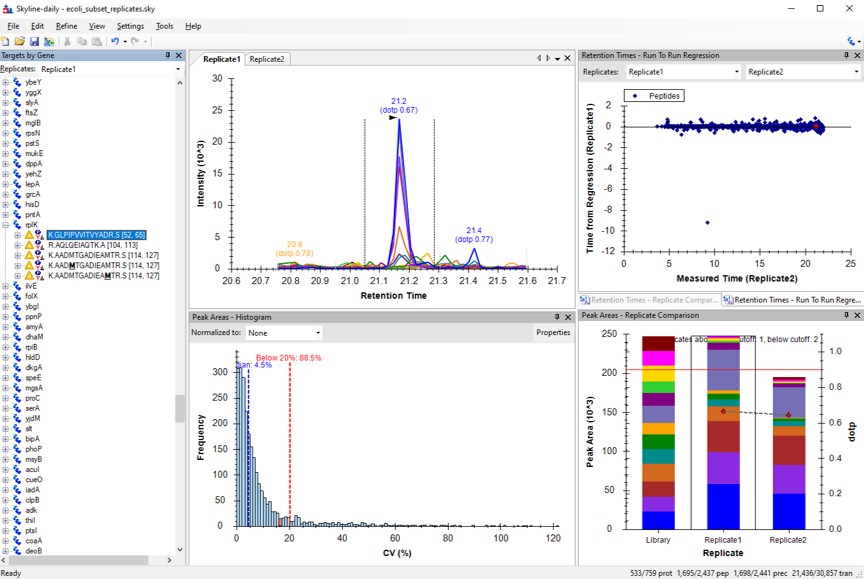
Final-Draft Creation
CV Refinement in Skyline
We have kind of a chicken-and-egg dilema now. As the .sky document currently has all of the up to 15 library transitions for each peptide, should we filter them first, and then filter the precursors by CV, or should we filter by CV and then filter the transitions? Here we opted to do the following to explore all the different options:
- Use Save As and save ecoli_subset_replicates_refined.sky. Sometimes when you save the document, the transitions go back to their original state. So it's better to save the document before you make any big changes.
- This switching of the transitions back to their original state happens if Settings / Transition Settings / Filter / Auto-select all matching transitions is selected. Go there and unselect that option, and press Okay.
- Run Tools / Thermo / PRM Conductor
- Select the Keep All Precs. option, so that we don't filter precursors, and keep a minimum of 3 transitions for each one. If there are only 2 good transitions, the 3rd one will be the next highest scoring, where the score is intensity x time_correlation.
- Press the Send to Skyline Button
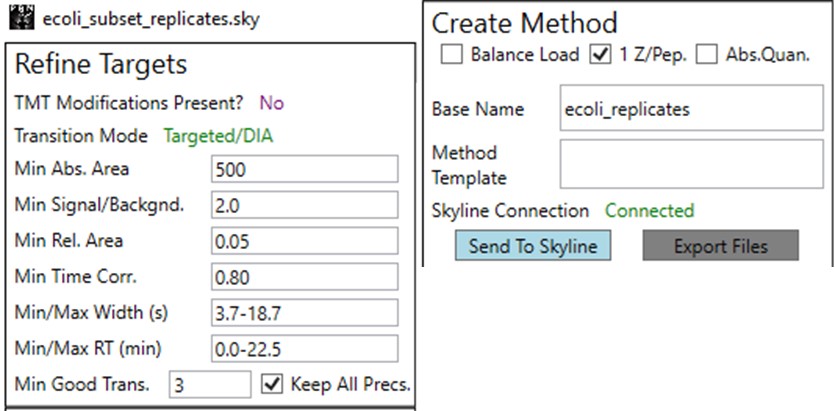
You can use the undo/redo buttons to see the effect that the filtering has had on the transitions. For example, in the figure below is shown the data for the AQLQEWIAQTK peptide before and after filtering.
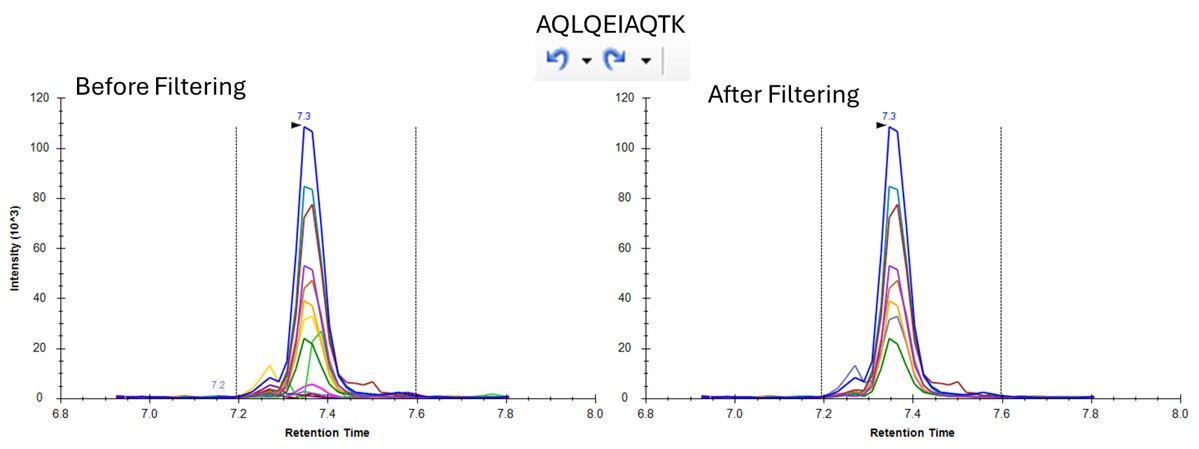
- One can click on the CV histogram, and Skyline opens a Find Results pane, that allows you to click on peptides and explore what the peaks look like at various CVs. You could also use View / Peak Areas / CV 2D Histogram to see a rather typical plot, where the most abundant peptides have the lowest CV's, in general, although bad CVs can be found at almost any intensity. Or good CVs at low intensity, for that matter.
Because we have a MS experiment in our method, the Total Ion Current normalization method is used, which can help normalize out experimental variation like autosampler loading amounts.
- Save the Skyline file as ecoli_subset_replicates_refined_cv.sky.
- Use Refine / Advanced / Consistency and enter 30 in the CV cutoff %. If you were going to use TIC Normalization, then be sure to select that option in the Normalize to: combo box. Press Okay. The CV histograms will update, and the number of proteins, peptides, precursors, and transitions will update in the bottom right hand corner of the Skyline document. You can use the undo/redo to check the effect. Save the Skyline document.
Filtering + Load Balancing
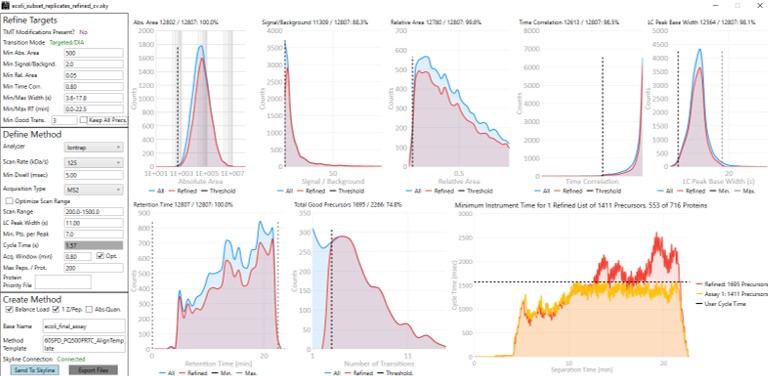
Now we will design the Final Draft assay. Our goal for this section is to ensure that the instrument can acquire quality data all the targets that are of interest. Here the experimenter can make certain concessions, such as, how many peptides per protein would give a usable result? Or am I willing to use 7 points across the peak instead of 10? We send the data to the PRM Conductor tool for analysis again using Tools/Thermo/PRM Conductor and get a View of the program like below. Compared to the earlier figure when PRM Conductor was run on the discovery results, these graphs are much different, because the precursors and transitions picked were already of such high quality. The percentage of transitions retained in the titles of each graph is close to 90% or greater. We set the LC Peak width to an even 11 seconds after hovering our mouse over the LC Peak Base Width graph to see the apex width, and set the Acquisition Window to 0.8 min.
This assay is what we would currently consider "normal", or conservative. There are 1411 precursors in a 24 minute method, which is about 3.5k precursors/hour. Had there been more precursors at earlier retention times, these settings would give about 5k precursors/hour. For fun, we have included HeLa results in the Step 1. DIA GPF\Processing\HeLaResults folder for those that wanted to explore what happens with a massive number of possible precursors.
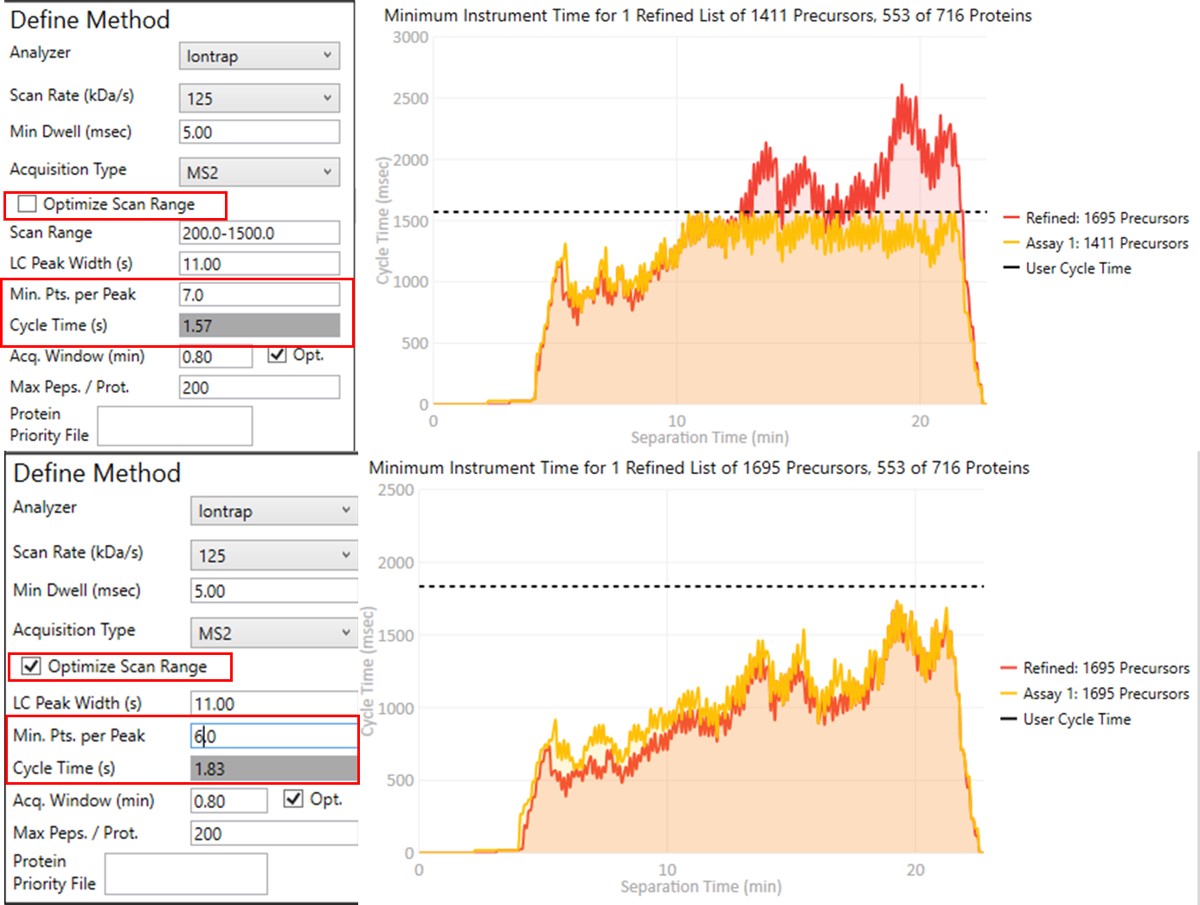
To realize the maximum throughput on Stellar MS, the user can select the Optimize Scan Range button, and could try using 6 points per peak, which is sometimes considered to be the Nyquist frequency for a Gaussian peak. We have successfully used such settings and observed little change in the results. The tradeoff being made with these settings is in the minimum amount of injection time per acquisition.
- The conservative mode guarantees about 13 milliseconds of injection time, because that's the largest injection time that doesn't slow down an HCD acquisition. If you made a method with the Maximum Injection Time mode set to Auto in the Method Editor, this is the injection time you would get. We like to use the Dynamic Maximum Injection Time Mode, where the precursors that have extra time in the their cycle, like the ones at 8 minutes, would get that extra time distributed to them.
- The aggressive mode only guarantees as much injection time as would not slow down the acquisition that has a customized scan range for the transitions that pass the PRM Conductor filters. This could be as little as the value in the Min Dwell Time box, currently set to 5 milliseconds. The rule-of-thumb is that signal-to-noise goes up (and LOQs get better) as the square root of the injection time.
- If you export a method with the Optimize Scan Range, you can see all the various, customized scan rates for each precursor.
- This mode can achieve more than 8000 peptides/hour throughputs. Play with the HeLa results to check this out.
Create Final-Draft Targeted Method and Skyline file
Notice that in above figures, the Balance Load check box is checked, and so the yellow Assay 1 trace in the Minimum Instrument Time plot is flattened up against the user Cycle Time. If the Max. Peps/Prot. value was reduced from 200 to 2, then the assay would contain only 869 precursors precursors, and there would be more space between the top of the Assay 1 trace and the horizontal cycle time line. Peptides are added from each protein in order of their quality from highest to lowest, as long as there is time at that point in the assay, and as long as the protein being considered has less than Max. Peps/Prot. peptides scheduled. It’s possible that an assay with fewer targets would have better quantitative performance, due to the longer available maximum injection times, and that could be of interest to the experimenter. In this example we leave the Max Peps./Prot. value at 200.
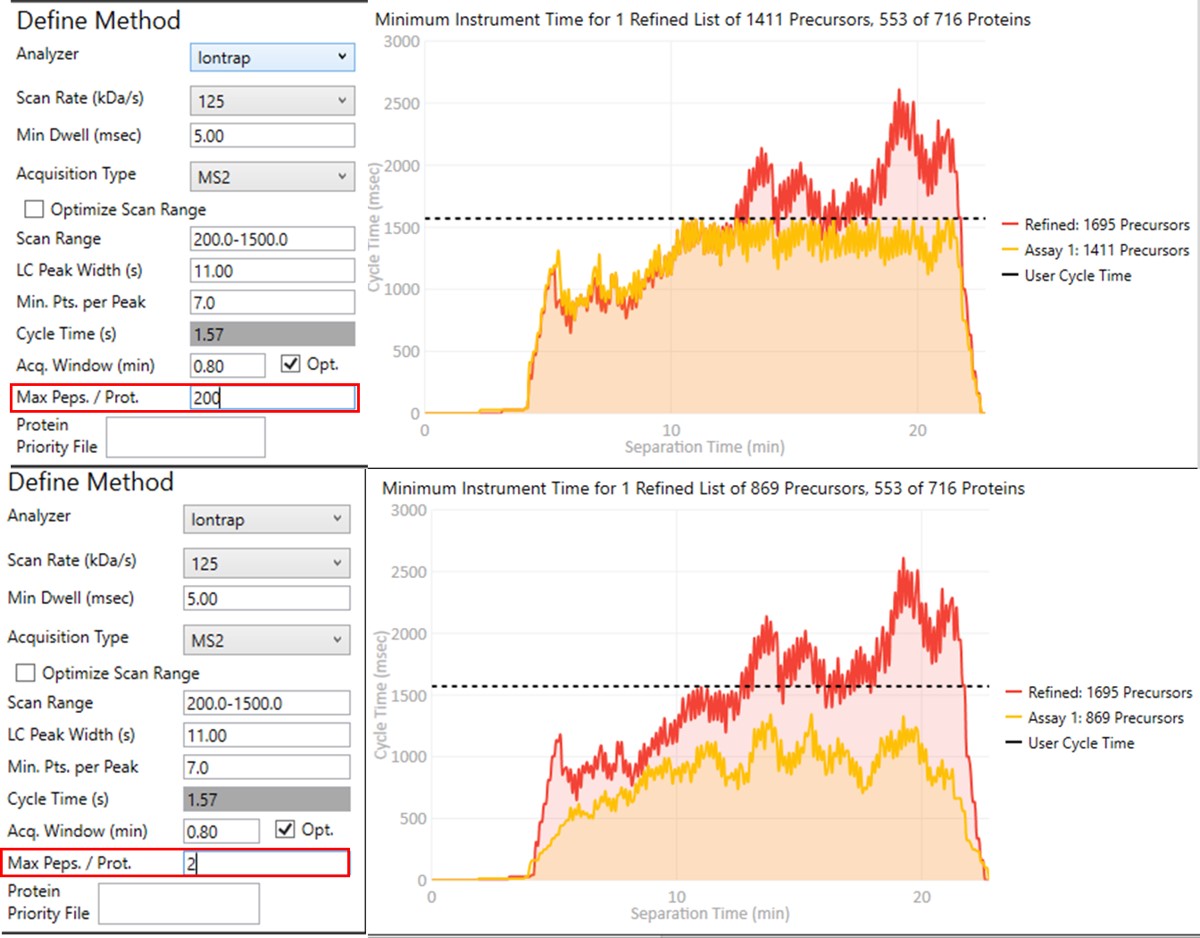
- Click on Export Files to export a method. Once again, some processing takes place as the raw files are analyzed, and finally isolation and precursor lists, as well as method and Skyline files are created. This is the final draft assay, ready to be used for targeted experiments.
- Normally we would use either the created Skyline file to collect our next data sets, or we would Save the current Skyline file with a new name, and press Send to Skyline from PRM Conductor to update it. In this case, because as mentioned above, the subset validation assays were slightly different when the data were collected compared to this walkthrough, we'll use the Skyline file actually used for data collection in the next step.
Analysis of Final Assay Replicates
Final Assay Replicates
Many researchers are not quite done once they have a "final-assay". Typically users want to perform a larger number of replicate injections with this candidate assay. A typical practice for assays that will be used for a large number of samples is to characterize the stability of the peptides and reproducibility of the LC/MS system using a 5x5 experiment. In this experiment, 5 different samples are digested and prepared, and each aliquoted into 5 vials, for 25 total. The vials are stored in the autosampler or a refridgerator at the same temperature, and each day for 5 days in a row, at least 5 replicates are acquired for each of the 5 vials. The inter-day, intra-day, and inter-sample variability can be assessed, and poorly performing peptides can be removed.
Here we will simply load some replicate data acquired with a final assay very similar to the one that we created in Step 2 of the walkthrough.
A Note about Spectral Libraries
An action that we could have included at the end of Step 2 is to create a final spectral library, once all the final peptides are refined and their retention times are known. In the future when replicates are imported, the retention time filtering for MS/MS IDs will be relative to the spectral library you have created, and not from the discovery runs, which can be useful. We can do this from the Step 2. Validation with Subsets\ecoli_subset_replicates_refined_cv.sky file.
- Use File / Export / Spectral Library
- You could use a name like ecoli_final_assay in the dialog that pops up. Note that when we exported a final method in the last step, the script actually created that library, so you don't have to overwrite it.
- To use the new library, you go to Settings / Peptide Settings / Library, and Edit list.
- Move the cursor to the bottom of the list, so the new library will be added there. Press Add.
- Find the location of the library you made, and give it a name.
- Then unselect the previous library, and select the new library.
- Press Okay to exit. Now the new library is being used.
- As it is, we'll use the library that comes with the file Step 3. Final Draft Replicates\ecoli_large_replicates.sky. Open this file.
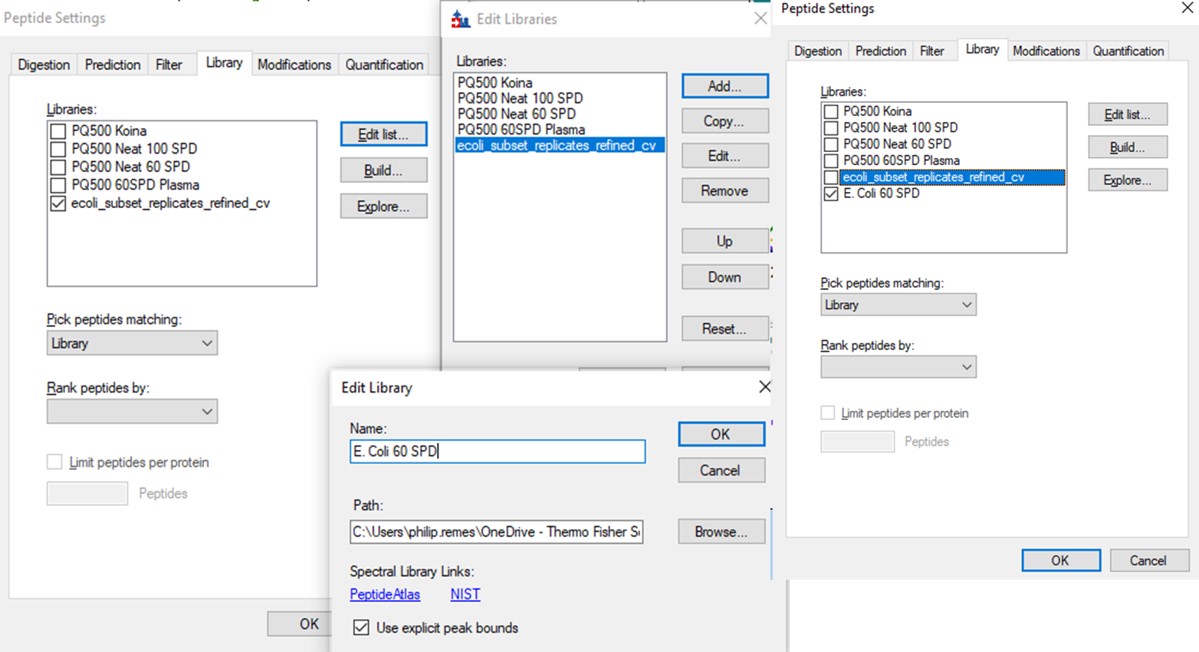
Loading and Analyzing the Results
- Use File / Import / Add single-injection replicates in files, and add the 8 files in Step 3. Final Draft Replicates\Raw.
- After these load, you can arrange the Skyline document as before.
- Save the Skyline file as ecoli_large_replicates_loaded.sky.
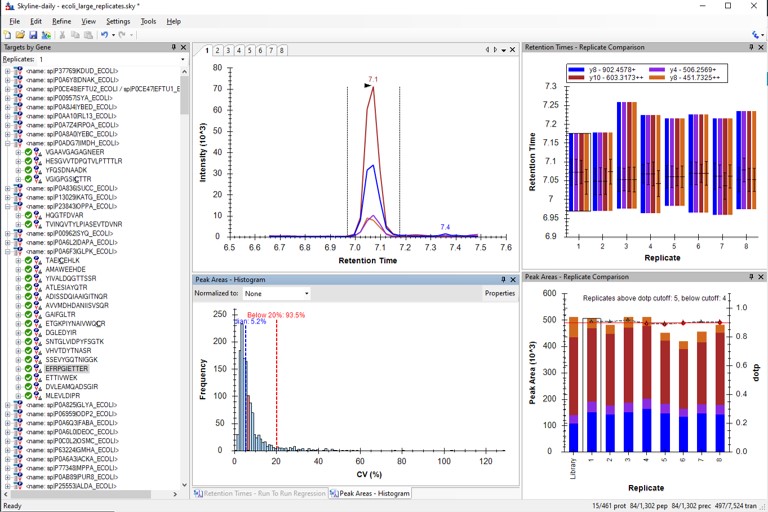
Use View / Retention Times / Regression / Run-to-Run. There are some peaks that are not consistently picked. Click on any of these outliers to see what they look like. Most are some kind of noise. Maybe they were "reproducible noise" in a previous step, or maybe there are interferences that get picked up over time with multiple replicates.
We could filter out the remaining peptides with poor CV's. This actually gets rid of all but a few of the retention time outliers. Additionally we could run PRM Conductor, and set some more stringent filtering settings, like in the figure below. We would unclick the Balance Load box to ensure that no precursors are filtered based on whether they fit in less than the cycle time, and press Send to Skyline.
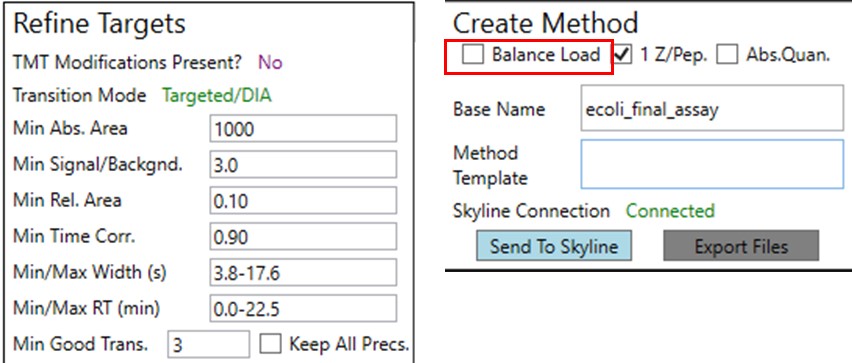
Here we see the effect on the retention time outliers from run to run. Almost all the outliers are gone.
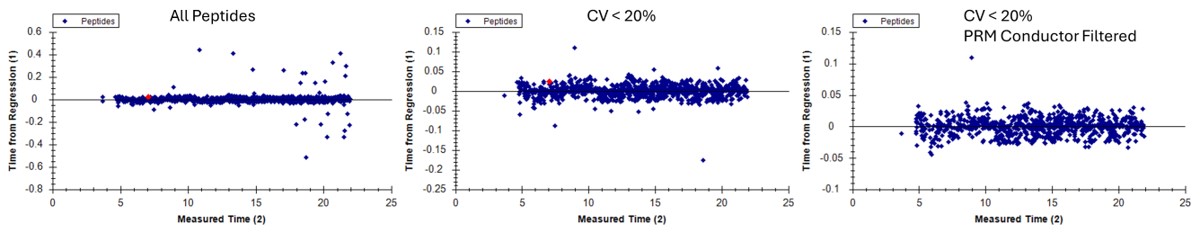
- Make sure that Settings / Transitions Setttings / Filter / Auto-select all matching transitions is not selected, and then Save As ecoli_large_replicates_loaded_refined.sky. This assay is has removed a few precursors, and could be used going forward.
Filtering Retention Time Outliers in Another Way
Let's try another way of filtering the retention time outliers. Go back and open the file Step 2. Validation with Subsets\ecoli_subset_replicates_refined_cv.sky.
- Open Settings / Peptide Settings / Prediction.
- Select Add from the Retention time predictor drop down menu.
- Select Add from the Calculator drop down menu.
- In the iRT standards drop down, select Add.
- In the Calibrate iRT Calculator, press Use Results, and enter a number like 100 in the Add Standard Peptides dialog that comes up, and press Okay.
- Give the Calculator a name like E. Coli PRM Assay and press OKay in the Calibrate iRT Calculator.
- In the Edit iRT Calculator, press Add Results, and then Press okay when asked to add these peptides, and if the standards should be updated.
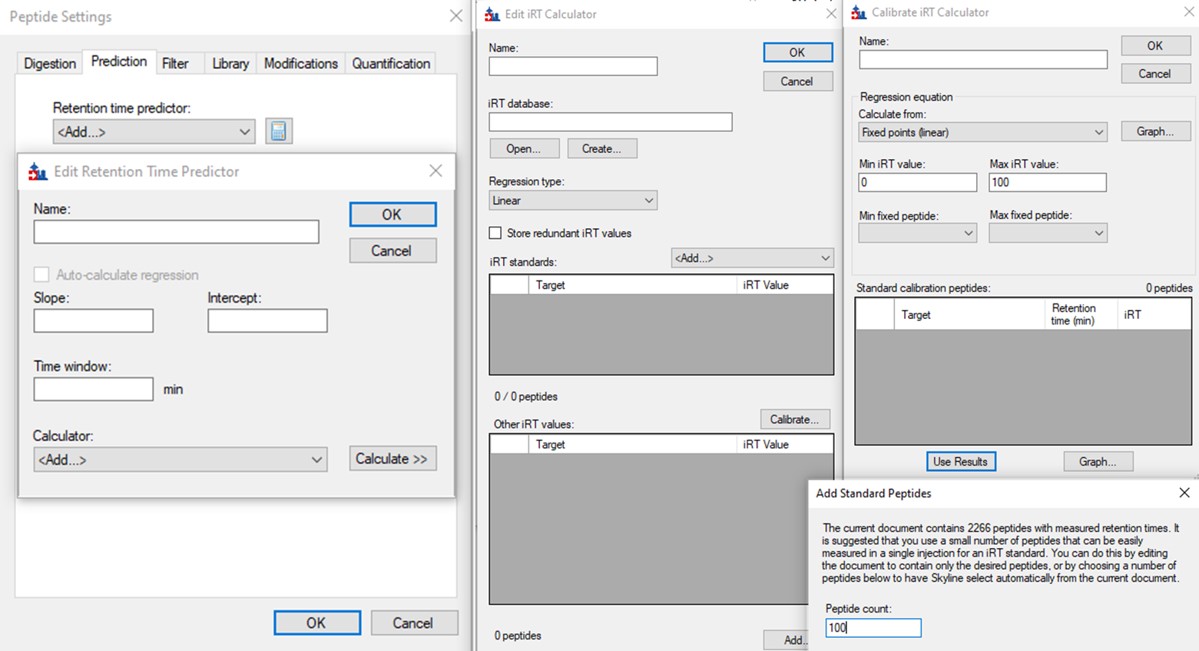
- Press Create on the iRT database button, and select Yes when ased to create a new database file. Save as Step 3. Final Draft Replicates/ecoli_irt_prm.irtdb.
- In the Edit iRT Calculator dialog still, give it a name like E. Coli PRM and press Okay.
- In the Edit Retention Time Predictor, set 2 minutes, and press Okay.
- We are still in the ecoli_subset_replicates_refined_cv.sky file, so we don't need to use the calculator here. Set None in the Retention time predictor, press Cancel, and close this Skyline file.
- Open up our ecoli_large_replicates_loaded.sky file, where we have all the retention time outliers.
- Go to Settings / Peptide Settings / Prediction and the E. Coli PRM name should be there. If it wasn't, for some reason, you could add the calculator we created from the Edit List option. Select the E. Coli PRM calculator.
- We were asked to add some standard peptides, because some of the ones that went into the calculator are not present. You can add them or not add them.
The background of the chromatogram plots is now beige, and there is a faint vertical line that says "Predicted" on the plots, like below.
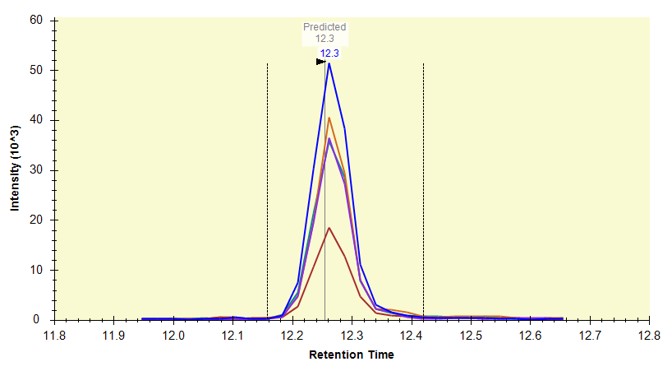
Use View / Retention Times / Regression / Score-to-Run, and on the plot that comes up, right click and in the Calculator menu, select the E. Coli PRM. Make sure that you are in the right-click, Plot / Residuals mode. Some of the dots in the plot are pink just because our document here has some peptides that weren't in the calculator. That's fine for this walkthrough.
- Use Refine / Advanced / Results and set Retention time outliers to 0.999 and press Okay. These peptides that were pink dots are now removed.
- You can do the same thing by right clicking the plot and setting the threshold, and then right clicking again and selecting Remove outliers.
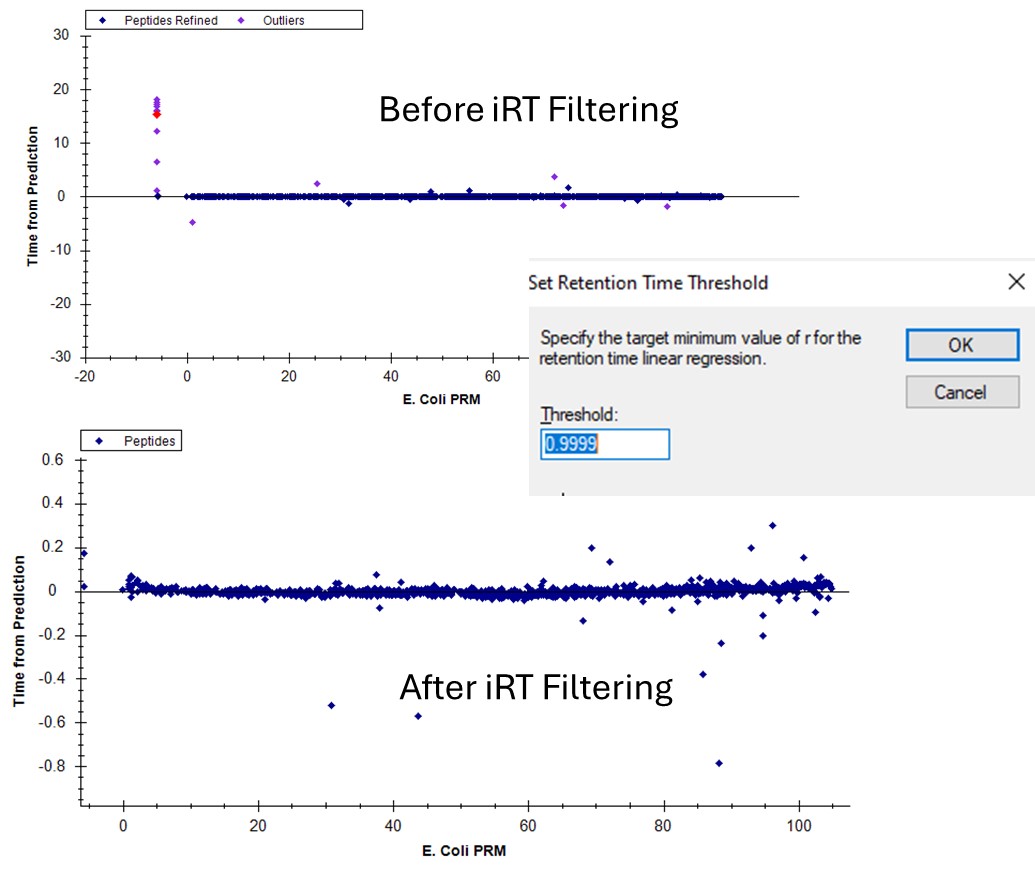
If we go back to the Run-to-Run regression, there are still a few outliers. Apparently we can't yet filter based on the experimental Run-to-Run deviations, but maybe in the future. This iRT filtering maybe wasn't as powerful in this example as just using the CV and PRM Conductor filtering, but it's another tool in our belt.
You've reached the end of this tutorial. Hopefully have a good idea of how to create an assay based on Stellar MS discovery results.