WELCOME TO THE LINCS PCCSE PANORAMA REPOSITORY
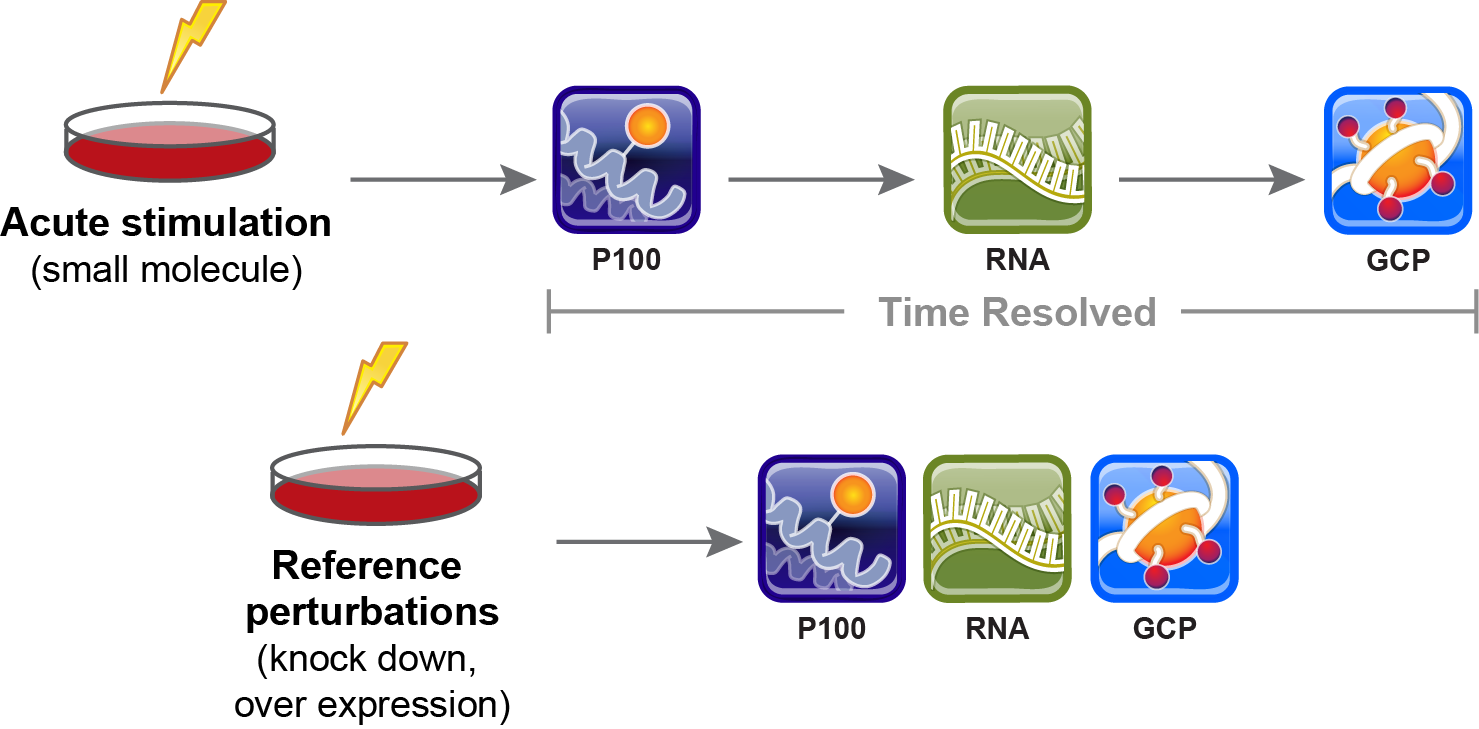
This is the LINCS Proteomic Characterization Center for Signaling and Epigenetics (PCCSE) Panorama Repository for the reduced representation phosphoproteomics assay ("P100") and global chromatin profiling assay ("GCP") primary mass chromatogram data. Our center makes systematic perturbations (drug treatments, gene disruption/editing, and directed differentiation) in a variety of biological models including cancer and neurodevelopment. We read out molecular changes in the cells in the spaces of phosphosignaling (P100) and histone modification (GCP). In collaboration with the LINCS Center for Transcriptomics, we also read out changes in transcription using their L1000 assay. Look here for more information about our center or download our poster here.
| PCCSE EXPERIMENTAL PARADIGMS | ANNOUNCING THE PCCSE PROTEOMICS CONNECTIVITY HUB |
 |
Go beyond the heatmaps! Explore connections and relationships among our perturbations in the Proteomics Connectivity Hub, a collaboration with the Connectivity Map Project. Free account creation required. |
Quick Links (Levels 1,2,4 data):
| ALL P100 DATA | ALL GCP DATA |
Quick Level 4 Data Visualization (& Download):
Note: this is probably what you want! For rapid access to visualizations of data, use the table below. Clicking on any link will lead to a visualization of data that we have collected. Each cell is a plate ID number followed by links to the data where available. Links beginning with "P" point to P100 data, while links beginning with "G" point to GCP data. The numbers after the leading character describe length of compound treatment before samples were harvested. All P100 data is acquired in DIA unless otherwise marked.
| MCF7 | PC3 | A549 | A375 | YAPC | NPC | Astrocytes | |
| Epigenetically-active drugs | 15 P3/G24 | 18 P3/G24 | 19 P3/G24 | 17 P3/G24 | 30 P3/G24 | 16 P3/G24 | 64 P3/G24 |
| Neurodevelopmental drugs | 23 P3/G24 | 24 P3/G24 | 25 P3/G24 | 22 P3/G24 | 31 P3/G24 | 20 P3/G24 | 65 P3/G24 |
| Kinase/Pathway Inhibitors | 29 P3/P6/P24/G24 | 34 P3/G24 | 33 P3/G24 | 28 P3/G24 | 32 P3/G24 | 27 P3/G24 | 66 P3/G24 |
| Cardiotoxic drugs | 60 P3/G24 | 61 P3/G24 | 58 P3/G24 | 59 P3/G24 | 62 P3/G24 | 63 P3/G24 | 67 P3/G24 |
Click here for more information on LINCS PCCSE Data Levels.
LINCS PCCSE Data Processing and Levels
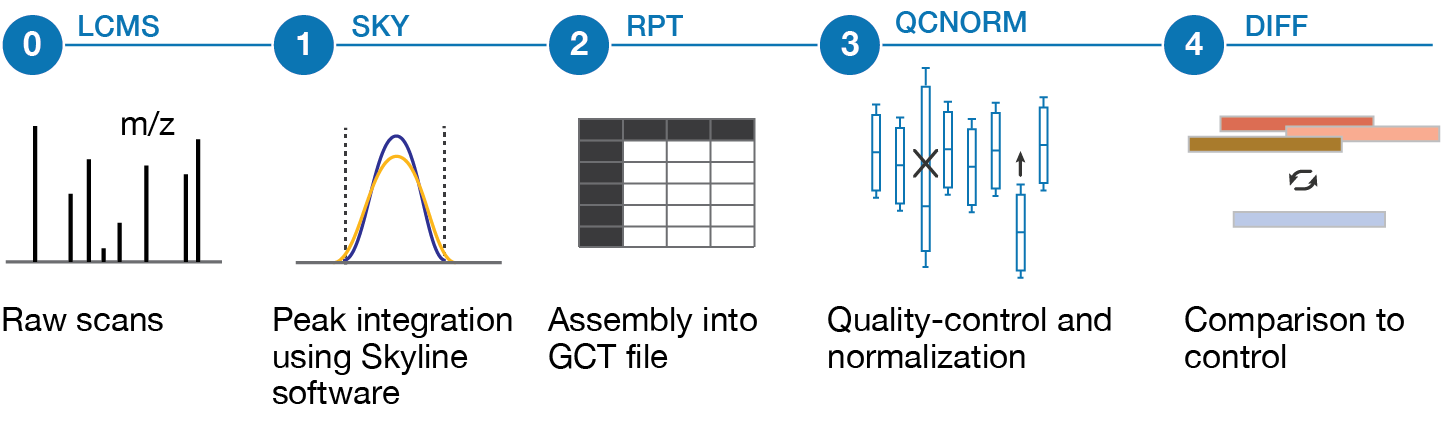
LINCS espouses the concept of making different data levels available for public use. Different data levels correspond different steps along our processing workflow. The LINCS PCCSE levels are defined as follows:
Level 0 - Raw Mass Spectrometry Data (LCMS) - will be available through a chorusproject.org repository in the future
Level 1 - Probe Reads (SKY) - Curated Skyline documents; available on this website, including metadata
Level 2 - Raw Numerical Data (RPT) - Matrix data of extracted signal ratios of endogenous probes vs. internal standards (log2 transformed); available on this website, including metadata
Level 3 - Normalized and QC'ed Numerical Data (QCNORM) - Matrix data derived from Level 2 after automated processing and normalization
Level 4 - Differential Quantification (DIFF) - Matrix data of Level 3 with plate-wide median ratio of each analyte subtracted from each sample; available on this website, including metadata
These data levels are mapped onto our processing pipeline as depicted below:
TIP: To browse and access our data sets, hover over the "LINCS" folder in the upper left corner: