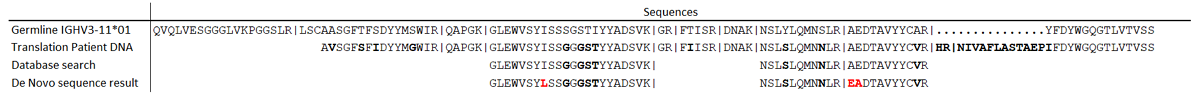Retrospective longitudinal monitoring of multiple myeloma patients by mass spectrometry using archived serum protein electrophoresis gels and de novo sequence analysis
Noori S, Zajec M, Russcher H, Tintu AN, Broijl A, Jacobs JFM, Luider TM, de Rijke YB, vanDuijn MM. Retrospective Longitudinal Monitoring of Multiple Myeloma Patients by Mass Spectrometry Using Archived Serum Protein Electrophoresis Gels and De Novo Sequence Analysis. Hemasphere. 2022 Aug 2;6(8):e758. doi: 10.1097/HS9.0000000000000758. PMID: 35935609; PMCID: PMC9348860.
M-proteins are antibody molecules produced by a clone of Multiple Myeloma cells. Thus, they are used as a disease activity marker. Traditionally, serum protein electrophoresis gels (SPEP) are used to quantify M-proteins. While effective for diagnostics, sensitivity of SPEP is limited. Here, we demonstrate that M-protein gels can be excised from SPEP gels, even when the band is undetectable by visible inspection. Clonotypic peptides are identified by de novo sequencing of mass spectrometry data, and these clonotypic peptides are then monitored in the longitudinal data covering the course of disease. Thus, it is possible to follow disease activity in a blood sample with a sensitivity that may be similar to that of minimal residual diseas platforms that rely on bone marrow biopsies.
Patient material
Materials used in this study are diagnostic SPEP gels (Hydragel, Sebia) from MM patients, archived at the Department of Clinical Chemistry, Erasmus MC. For SPEP analysis, MM patient serum has been processed and used for diagnostic purposes. The archived SPEP gels were stored as dried agarose gels supported by a plastic carrier at room temperature.
MM patients (n=9) were retrospectively selected based on the data in the hospital information system. Patients were selected to have SPEP detectable M-protein at diagnosis (>1 g/L), and at least one period when the M-protein was absent by SPEP and IFE, followed by again detectable M-protein. From these patients M-protein DNA/RNA sequence was not available. In the Supplemental Digital Content the available clinical data has been listed for the selected patients including SPEP, free light chain, urine electrophoresis, bone marrow MRD data (10-4 flow cytometry) as well as the main treatments. Patient samples and clinical data were coded as specified in the Dutch code of conduct for biomedical research (institutional review board approval MEC-2019-0342). Gel samples from retrospectively selected patients were measured in SPEP gels generated from 2010 to 2016, and all available gel samples have been measured by MS. Patient information is provided in the data repository.
A dilution series was prepared from a sample (16.6 g/L of M-protein by SPEP) of a patient that also has DNA sequence data on the M-protein heavy chain. The dilutions were analyzed using SPEP to validate the de novo sequencing approach and the quantitative performance of our method.
Excision and trypsin digestion of the M-protein bands
Cutting the M-protein bands was performed in steps to ensure that the correct area of the gamma/beta fraction was cut even when the M-protein band was not visible. The width of the M-protein band and the distance to the albumin band were measured. At time points when M-protein was not detectable by SPEP, these metrics were used to locate the proper position in the gel. With a scalpel the M-protein band was excised from the gels. The dilution series of the reference patient was cut as described previously.
Each M-protein band was digested separately in a 1.5 mL Eppendorf tube, all volumes were 60 µL unless specified otherwise. First, the bands were washed with water, 50% acetonitrile (ACN), 100% ACN and 50% ACN in 100 mM ammonium bicarbonate (ABC), then dried in Savant SC210A SpeedVac concentrator (Thermo Fisher Scientific, Munich, Germany) for 5 minutes. Reduction and alkylation were performed using 10 mM dithiothreitol at 56°C for 45 minutes, and 55 mM iodoacetamide at room temperature in the dark for 30 minutes. Afterwards, gel bands were washed with water, 50% ACN, 100% ACN and 50% ACN in 100 mM ABC and dried. Then, 0.1 % RapiGest SF (Waters, Milford, MA) in 50 mM ABC was added for 10 minutes at 37 °C. After drying, 600 ng of gold-grade trypsin (Promega, Madison, WI) in 50 mM ABC was added and incubated at 4°C for 5 minutes. The solution was collected and 50 mM ABC was added, followed by incubation at 37°C overnight. Tryptic peptides were extracted once with 1% trifluoroacetic acid (TFA) and twice with 0.1% TFA in 50% ACN. All extracts and trypsin solution were combined and completely dried before resuspension in 25 µL 0.1% TFA. All samples were cleaned with C18 ZipTips (Millipore, Burlington, MA).
Liquid chromatography-mass spectrometry (LC-MS)
Liquid chromatography was carried out on a nano-LC system (Ultimate 3000, Thermo Fisher Scientific, Munich, Germany). Sample volumes of 10 µL were injected and separated on a C18 column (Acclaim PepMap, 75 µm ID × 250 mm, 2 µm, 100 Å ; Thermo Fisher Scientific, Munich, Germany). A 30 minute gradient of 4%–38% was used; A: 0.1% formic acid in water; B: 80% acetonitrile, 0.08% formic acid in water.
DDA MS measurements
Data Dependent Acquisition (DDA, Shotgun) measurements were performed on a QExactive HF Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). MS1 scans (375.00–1500.00 m/z) had a resolution of 60,000, AGC target 3e6, maximum injection time 60 ms. MS/MS spectra used HCD fragmentation at 28% normalized collision energy on the top 20 precursors at resolution 15,000, dynamic exclusion 40 s, AGC target 5e5, maximum injection time 50 ms and isolation width 1.4 m/z.
Targeted MS measurements
Targeted MS/MS spectra used HCD fragmentation at 27% normalized collision energy, resolution 30,000, AGC target 5e5, maximum injection time 250 ms and isolation width 1.4 m/z. All peptides are listed in Supplemental Table S-2 as used for quantitation.
De novo sequencing of the mass spectra
De novo sequencing was performed on DDA as well as targeted MS raw data files using Peaks Studio 6.0 (Bioinformatics Solutions Inc., Waterloo, Canada). The following settings were used: enzyme trypsin, Orbitrap instrument, HCD fragmentation, methionine oxidation and carbamidomethylation were selected as variable and fixed post-translational modifications, respectively. Parent ion tolerance of 5 ppm and fragment ion tolerance of 0.02 Da were allowed. Where targeted and DDA MS data resulted in de novo sequences with a difference, those based on the targeted data were used due to the superior signal in these.
Selection of patient-specific M-protein peptides from de novo sequencing data
Of each patient, the sample of the first timepoint was measured in order to select patient-specific M-protein peptides. Peptide candidates had to fulfill several criteria. First, a patient-specific M-protein peptide candidate has to be highly abundant in the patient of interest and absent or a minimal signal (<1%) in all other patient samples. Progenesis QI (version 2.0, Waters, Milford, MA) was used for label-free quantitation of MS data and combined with de novo sequencing information. Other selection criteria were: the peptide has an de novo sequencing total local confidence (TLC) above 7; the peptide should not be identical to an immunoglobulin germline sequence (IMGT), ensuring that the sequence contains mutations that can make it specific to the patient. Also, a search with IgBlast (www.ncbi.nlm.nih.gov/igblast/) was used to show peptide homology to immunoglobulins, which was >68% for all selected peptides.
Longitudinal M-protein monitoring
Patient-specific M-protein peptides were used to monitor the disease course of each patient. All samples were measured with targeted MS data analyzed in Skyline. A spectrum library was created, and samples with a library dot product exceeding 0.8 were considered positive. The area of the peaks were plotted with other clinical information available for these patients. As the clinical records were inconsistent in reporting SPEP data below 5 g/L numerically, all numeric data between 1 and 5 g/L were plotted as 5 g/L. Samples shown as negative for M-protein were assessed as such by a laboratory specialist based on the combined data from SPEP, IFE and nephelometry that was available at the original time of analysis.