Table of Contents |
guest 2025-09-16 |
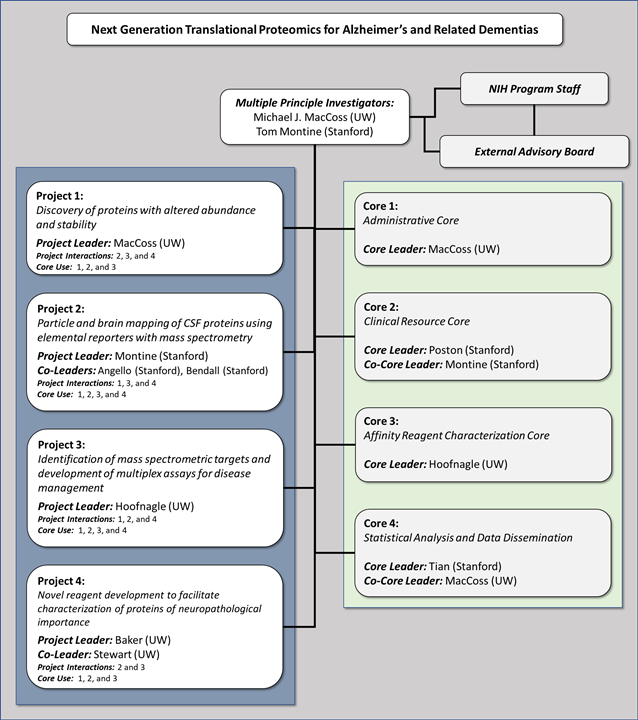
Project 1: Discovery of proteins with altered abundance and stability
Project 2: Particle and brain mapping of CSF proteins
Project 3: Development of mass spectrometry assays for disease management
Project 4: Novel reagent development to facilitate characterization of proteins of neuropathological importance
Core 1: Administrative Core
Core 2: Clinical Resource Core
Core 3: Affinity Reagent Characterization Core
Core 4: Statistical Analysis and Data Dissemination
Next Generation Translational Proteomics of Alzheimer's Disease
Overview
Alzheimer's disease (AD) is a major, growing global public health problem. This is a daunting scientific challenge and solutions will come only from innovative research. We have brought together a unique interdisciplinary team of investigators with the goal of bridging the divide between state-of-the-art technologies and translational applications.
Our U19 research program will consist of four projects and four cores that are synergistic to our mission. Moreover, our research team is uniquely suited to the development, validation and translational application of new biomolecular assays to reflect AD pathophysiology.
Part of the project? Click HERE for internal resources (agendas, meeting logs, etc).
Learn more about each project or core by clicking on the icons below!
|
Project 1
|
Discovery of proteins with altered abundance and stability.
|
Project 2
|
Highly multiplexed particle and brain mapping of CSF proteins.
|
|
Project 3
|
Identification of mass spectrometric targets and development of multiplex assays for disease management.
|
Project 4
|
Novel reagent development to facilitate characterization of proteins of neuropathological importance.
|
|
Core 1
|
Administrative Core |
Core 2 |
Clinical Resource Core |
|
Core 3 |
Affinity Reagent Characterization Core |
Core 4 |
Statistical Analysis and Data Dissemination Core |
Project 1: Discovery of proteins with altered abundance and stability
Despite the association between the levels of CSF Aβ42, tau, phosphorylated tau and underlying AD pathology, measures of biomarker accuracy for clinical diagnosis vary widely between studies. Given that other neurodegenerative conditions can present with AD-like clinical symptoms, and individuals with AD frequently have comorbid pathologies, additional markers are needed that can aid in differential diagnosis and identify mixed pathologies. Over the last decade, many candidate biomarkers have been identified, reflecting a range of pathophysiological processes including cholesterol metabolism, neuroinflammation and amyloid processing. However, few, have been adopted in clinical practice or been validated in large independent cohorts.
The MacCoss lab and others have been pioneering the development of next generation proteomics methods as an alternative to the classic stochastic mass spectrometry-based methods. These new methods offer a hybrid between a targeted and global proteomics strategy. While mass spectrometry data is collected in an unbiased way, the data is analyzed in a targeted strategy where specific peptides, albeit 1000s are analyzed using prior information. Thus, the reproducible targeting, throughput, and confident MS/MS-based quantification of parallel reaction monitoring (PRM) can be combined with classical discovery methods’ ability to qualitatively detect thousands of proteins. These new methods based on systematically collected mass spectrometry data can offer similar quantitative figures of merit and can be validated in analogous fashion to clinical assays.
Despite advances in proteomics technologies, most attempts at discovering new CSF markers have either used 1) stochastic sampling methods (e.g. data dependent acquisition) with poorly characterized quantitative performance, 2) small sample cohorts, 3) focused entirely on total CSF, 4) ignored protein processing, and 5) did not consider protein misfolding or stability. The purpose of this project within the cooperative research program is to take our methods to another level – apply true quantitative methods to large well characterized cohorts and extend them to functionally relevant subpopulations.
We have a CSF assay that can measure >1050 proteins from completely unfractionated material with figures of merit of within/between day precision, linearity, LOD/LOQ, etc... Furthermore, we can use this assay on subsets of the CSF proteome to assess functionally relevant aspects of the neurobiology including quantity as part of solution or CSF particles, intact protein MW, and protein stability. Finally, we have enough throughput to apply these analyses on a scale sufficient to eliminate the chance that an observation is due to an aberrant subpopulation.
Project 2: Particle and brain mapping of CSF proteins
Alzheimer’s disease (AD) is a major threat to health of older individuals and is a looming public health disaster. Promised solutions in diagnostics and therapeutics will come only through research. Project 2proposes to gain biological and medical insights into the proteins discovered in the Center by multiplex mapping to specific CSF extracellular vesicles (EV) and lipoproteins (LP), and to specific cells and subcellular structures in specific regions of brain.
Proteins in CSF remain the best performing biomarkers for AD. CSF proteins exist in various forms: free in solution, on a unique class of LP, and on EV that include exosomes, microvesicles, and apoptotic bodies. LP and EV have varying mechanisms of production and biological functions. Localization and relative quantification of proteins of interest discovered in Project 1 to each class of CSF particles will provide powerful insight into cell of origin, cellular biology, and potential medical meaning.
Further biological and medical insights can be gained by carefully mapping proteins in human brain. Indeed, the power of multiplexed imaging is its ability to reveal co-localization or mutual exclusivity, and to infer regulatory roles and gain mechanistic insight. For example, we think very differently about the biological roles of proteins restricted in their expression to synapses vs. mitochondria, glutamatergic vs. GABA-ergic neurons, or hippocampal pyramidal neurons vs. striatal medium spiny neurons. In pathologic states, co-localization with hallmark pathologic structures (major protein) highlights pathways of injury and response to injury: senile plaques (amyloid beta peptides), neurofibrillary degeneration (paired helical filament-tau), Lewy bodies and neurites (phospho-alpha-synuclein), and phospho-TDP-43 inclusions.
We will test the hypothesis that multiplex analysis of CSF particles as well as subcellular, cellular, and regional mapping will provide key biological and medical insights into CSF proteins discovered by de novo proteomic analysis of CSF in Project 1. The same probes will be applied to CSF LP and EV using flow cytometry, and to brain regions using Multiplexed Ion Beam Imaging (MIBI).
Project 3: Development of mass spectrometry assays for disease management
The pathological hallmarks of Alzheimer’s disease are the neurofibrillary tangles and amyloid plaques that form in gray matter regions of the cerebral cortex. Misfolded and aggregated proteins reside within those pathological features, which may to be central to the neuronal death that causes regional atrophy, dementia, and ultimately death. The cerebrospinal fluid that bathes the central nervous system is the fluid most proximal to the disease and has been the focus of biomarker discovery for more than 30 years. There are two important gene products that are present in cerebrospinal fluid and have been shown to be predictive of disease activity and cognitive decline in patients that present with mild cognitive impairment: (1) the amyloid precursor protein, which gives rise to Aβ(1-42) and other fragments, and (2) tau, which has many phosphorylation sites. The deposition of Aβ(1-42) as amyloid aggregates appears to lead to the death of adjacent neurons. The resulting injury increases the amount of tau protein released into the cerebrospinal fluid. From previous biomarker studies of Aβ proteins and tau, it seems likely that the entire disease process, from Aβ(1-42) deposition to cell death, is orchestrated differently than in other neurodegenerative diseases. As a result, it is a compelling hypothesis that novel biomarkers, which may be mediators of disease, are present in the cerebrospinal fluid and could add more information over the current biomarkers used in the diagnosis of Alzheimer’s disease.
There have been previous attempts to use discovery proteomics to identify proteins that are differentially expressed in cerebrospinal fluid in patients with Alzheimer’s disease compared with controls and there have been few efforts to expand upon these studies in a clinically meaningful way. However, some of these efforts have identified the presence of additional fragments of amyloid precursor protein [i.e., in addition to Aβ(1-42)] and other proteins, which suggests that post-translational changes may be important in the formation of amyloid, the dysfunction of neuronal synapses, and the subsequent death of neurons.
In this project entitled, “Identification of mass spectrometric targets and development of multiplex assays for disease management,” our objective is to identify the post-translational modifications that lead to altered LP/EV concentration, molecular weight, and stability in cerebrospinal fluid. Part of the reason that the clinical translation of Aβ(1-42), tau, and phosphorylated tau as cerebrospinal fluid biomarkers took decades was that reproducible assays were not available early on. As a result, another overarching goal of this project is to develop precise, transferable, validated targeted proteomic assays to quantify proteins in cerebrospinal fluid that can be used to investigate disease mechanism and predict poor outcomes.
Project 4: Novel reagent development to facilitate characterization of proteins of neuropathological importance
Project 4 will test the hypothesis that computationally designed protein “minibinders” and logic-gated switches targeting cerebrospinal fluid (CSF) protein biomarkers and protein particles can serve as versatile capture and detection agents to vastly improve the molecular characterization of CSF samples. The availability of these reagents should support the overall U19 goal of improving our understanding of CSF biomarkers as direct measures of age-related cognitive decline and Alzheimer’s Disease (AD) pathophysiology.
We have integrated our work plan within the highly focused U19 Project: Next Generation Translational Proteomics for Alzheimer’s and Related Dementias to test the above hypothesis. In collaboration with Projects 1–3, our main goal is to develop an optimized set of computationally designed minibinders (hyperstable binding proteins of length <65 aa) as CSF protein biomarker capture agents, and ultra-specific logic-gated reagents for detection of CSF particles that have two defined protein components.
The research will iterate between computational design and experimental testing, with feedback at each stage from CSF assay experiments conducted in collaboration with Projects 1-3 which will guide improvement of the minibinder design methods for capture of specified CSF biomarker protein targets, and for ultra-specific logic-gated detection of CSF particles with two composite protein components. The outcomes will be (i) specific CSF biomarker capture and detection systems for AD and other age-related neurodegenerative disorders, and (ii) an integrated computational-experimental pipeline for rapid on demand engineering of new protein based diagnostic agents for neurodegenerative disorders in general. Therefore, Project 4 relies on a close collaboration with Projects 1-3 and Cores 1-4 within the highly interactive U19 program.
Core 1: Administrative Core
The Administration of the U19 will build on strategies learned and used over the last 15 years of management of numerous interdisciplinary centers, program projects, consortia, and public/private research collaborations. The greatest strength of our interdisciplinary resource program is a highly experienced and diverse team of investigators who have worked together productively for a very long time. Our program is highly focused on the development and application of state-of-the-art technology. Our resource leverages a unique group of individuals who are in a strong position to bring these technologies forward into a series of deliverables to make a translational impact to monitor Alzheimer’s disease progression and/or response to treatment. We will continue to use the administrative capabilities we have learned and put in place over the last 15 years to ensure an effective and functioning resource for the scientific community.
Core 2: Clinical Resource Core
Alzheimer’s disease (AD) is a major threat to health of older individuals and is a looming public health disaster. Needed solutions in diagnostics and therapeutics will come only through research. Our contribution to this overall project is to provide clinical resources to support the Projects. This includes consensus clinical diagnosis, systematic neuropsychological test results, neuroimaging data, CSF obtained by research quality lumbar puncture, and brain autopsy.
Our Core will leverage existing clinical research infrastructure provided by the Stanford AD Center and the Stanford Udall Center. Additional samples also will be provided by our long-time collaborators, Dr. Doug Galasko from the UCSD AD Center, and Dr. Thomas Beach from Banner Research Institute. The Clinical Resource Core’s Specific Aims are: 1) Consensus Clinical Diagnosis and Neuropsychological Test Results, 2) CSF Samples and 3) Brain Autopsy
Core 3: Affinity Reagent Characterization Core
Affinity reagents are essential to almost any modern scientific investigation of human disease. Antibodies are the most widely used family of affinity reagents and when cloned and produced in culture represent a powerful tool that can be continually regenerated. Many monoclonal antibodies boast strong interactions, slow off-rates, and good specificity. To supplement monoclonal antibodies, the Baker laboratory at the University of Washington has developed techniques to design small proteins de novo that bind molecules of interest. The overall goal of this Core entitled, “Affinity Regent Core,” is to provide the reagents needed for Projects 2 and 3 by (1) developing new monoclonal antibodies to be used in different assays, (2) labeling antibodies and novel small protein affinity reagents for use in CyTOF and brain imaging, and (3) carefully characterizing each reagent to ensure that different batches of affinity reagents behave similarly.
Core 4: Statistical Analysis and Data Dissemination
The overall function of the Statistical Analysis and Data Dissemination (S2D) is to provide strong support to data management, data dissemination, study design, statistical analysis and manuscript writing for proposed research projects in the center for Translational Proteomics in Alzheimer’s Disease. High quality data is the cornerstone for the success of the center for Translational Proteomics in Alzheimer’s Disease, since almost all the center-supported research will be data driven. Therefore, it is extremely important to build efficient data management systems to house data of various types and analyze them appropriately for both generating important discoveries as well as guiding the conduction of the proposed research. The core team has rich experience in conducting Alzheimer’s Disease-related research and plan to focus on managing data, providing high quality statistical support throughout the research and working with laboratory scientists to aide them in their own analyses of the data.