Table of Contents |
guest 2025-07-01 |
Absolute Quantitation - PQ500
Label Free - E. Coli
Expert Review Walkthrough
Expert Review Introduction
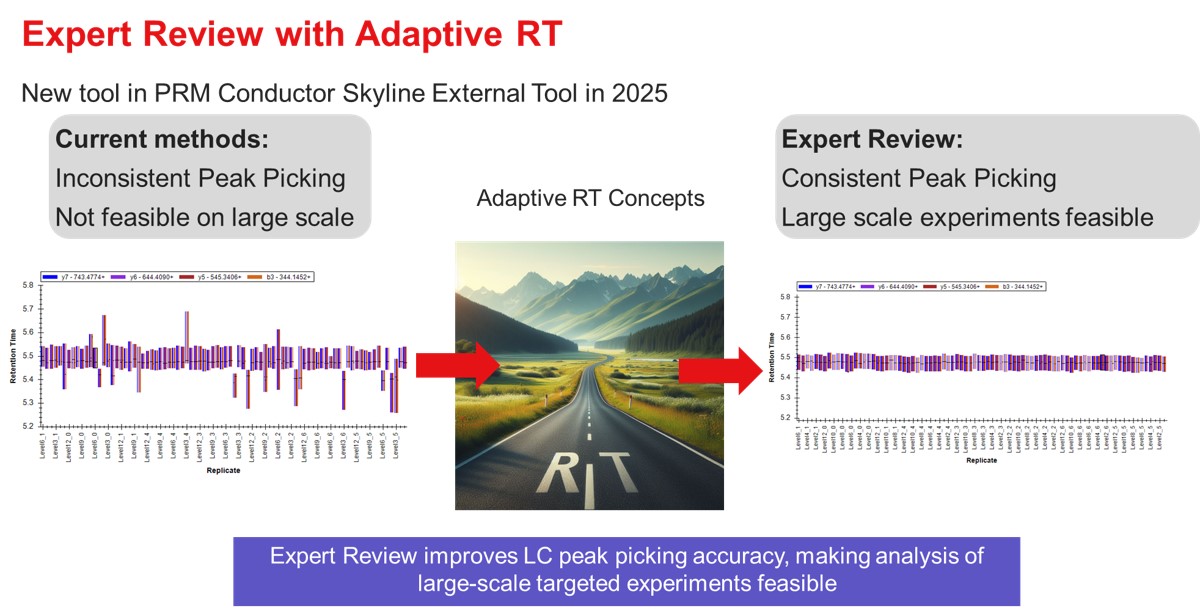
Targeted MS experiments have traditionally been of relatively smaller scale than discovery experiments,yet the data analysis can be prone to high rates of incorrect integration boundary determination. With the introduction of new MS hardware and acquisition strategies, notably Stellar MS with Adaptive RT and PRM Conductor, targeted experiments can routinely exceed 1000 targets. This increased scale necessitates improved tools for data analysis. Expert review is a software tool that improves the LC peak boundary imputation process, through the use of replicate-level and experiment-level analysis. The methodologies used were inspired by our algorithm used for real-time retention time alignment, known as Adaptive RT.
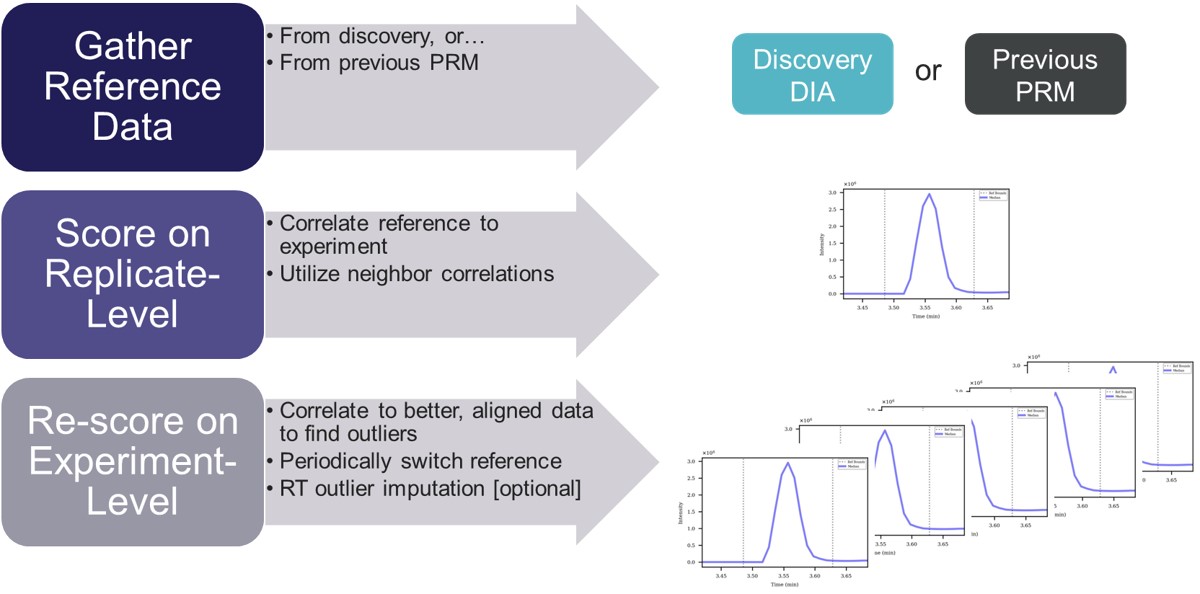
- To use Expert Review, you should have a reference set of data, where the all the peaks have been picked correctly. If you are doing a label-free experiment, the easiest way to obtain a reference file is to have a set of discovery data with compound IDs in it, ensuring that the correct peak was initially selected in Skyline. If the experiment you are doing uses heavy standards, then the reference data can come from injections of the neat standards.
- Expert Review utilizes two levels of analysis, a Replicate-level scoring based on an analysis of each replicate in isolation, and an Experiment-level re-scoring for each precursor, that considers all of its replicates.
Expert review has mostly been used for analysis of dilution curves, which are challenging test cases for integration boundary determination, because by design there are many replicates where the analytes of interest are diluted to below limits of detection. We have found via manual inspection in many cases that Expert Review makes no errors, and those errors that do happen are typically either very small, or caused by the use of the RT outlier correction, which is not suitable for abrupt RT changes, and is an option that can be turned off.
Absolute Quantitation - PQ500
Absolute Quantitation - PQ500
For this walkthrough, we will use data from our paper on Stellar MS with Adaptive RT and PRM Conductor. This section uses a dilution of the Biognosys PQ500 sample in plasma.
- Obtain the Skyline data files in this Panorama repo, in the Raw Data \ Expert Review folder. Open the Skyline file Absolute Quantitation \ 60spd_dilution_lightheavy_skyline.sky.
For me, the document opened with the NLVVIPK peptide selected, illustrating one of the great challenges of performing dilution curves with samples like the PQ500. You can use Ctrl+F and search for this peptide sequence, or peruse the document ande find many other such examples. Many of the targets, like this one, don't have appreciable endogenous signal, therefore peptides like this one will be problematic to analyze if the Skyline document is set to Settings / Peptide Settings / Modification Settings / Internal Standard Type / Light. Of course there are some peptides. like LVTDLTK from Albumen, that have enormous endogenous signal, and for which there is no issue using the Light Internal Standard Type for a dilution curve. If the Internal Standard Type is set to Heavy, there traditionally were problems when the dilutions are low enough that there is little to no Heavy standard signal, and the boundaries would get set more or less randomly. Expert Review aims to fix this problem.
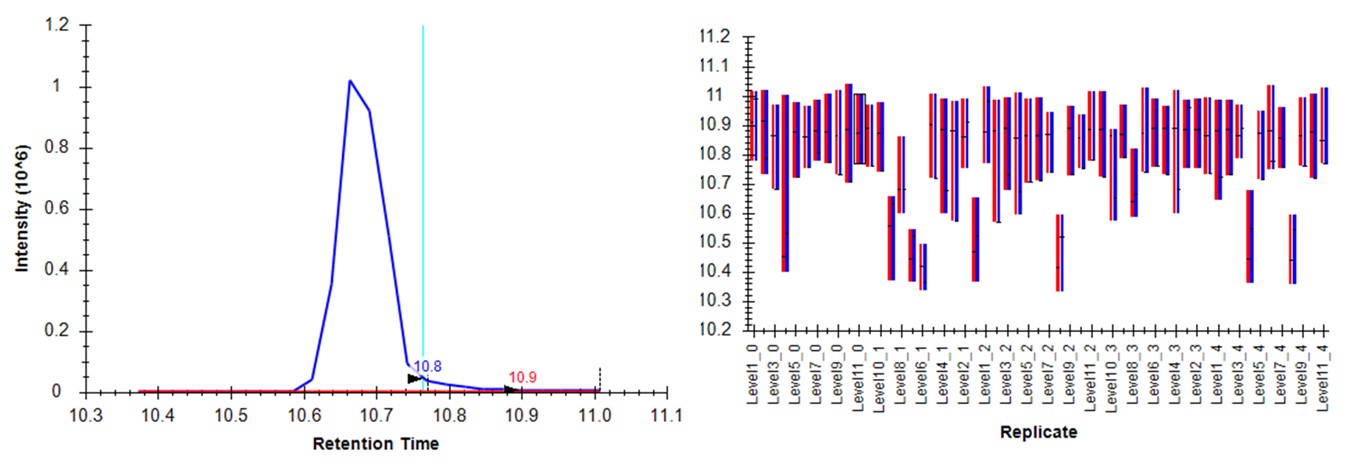
Expert Review - Light Boundary Mode
First we will look at the Expert Review results with the light Isotope boundary type, to illustrate the potential pitfalls of this mode for large assays that don't have much endogenous signal for many targets. Launch Expert Review after having installed or upgraded your PRM Conductor instance, by running Tools / Thermo / Expert Review. You will see the green progress bar move as Skyline exports the data set, and finally Expert Review will open. If this is the first time that you've run Expert Review, the Reference Grid will look like below, where the replicates for this experiment are listed. The replicates are listed from first-acquired to last-acquired, and the first replicate has a checkbox next to it.
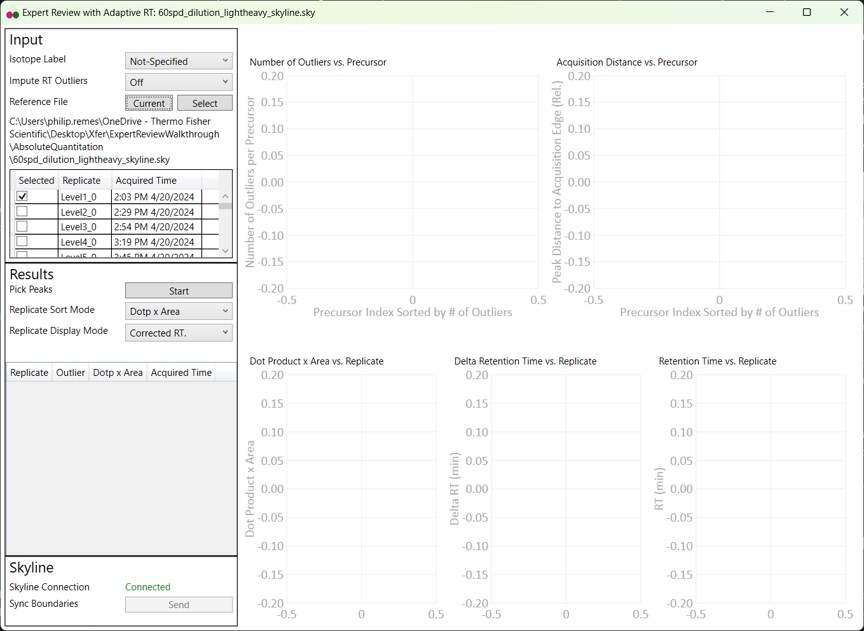
- If you select the checkbox, the grid will be highlighted in red, and you can't press the Results:Start button. We always need to have at least one reference replicate selected.
- If you are using a replicate from the same Skyline file you are using for analysis, and it isn't the first-acquired replicate, you'll have deselect the first replicate, find the replicate you want, and select it.
- I like to save the reference replicate in another separate Skyline file and press Select to open a file-chooser, and select that file.
- Expert Review remembers the last reference file that you used, which is someties convenient, but sometimes annoying when you switch experiments and forget to change the reference, and then there will be errors when your reference precursors don't include your experiment precursors.
- Press the Select button, and choose the other Skyline file in the Absolute Quantitation folder, P1_Neo_60SPD_HeavyLight_Reference.sky. This file has the last replicate from a test of the measurement precision of the assay.
- For the first test, select Isotope Label Light. When Isotope Label is set to Light or Heavy, a new option appears, Use Other Label's Info. This allows to use the data from the other isotope label, in this case the heavy data, to ensure a more consistent integration boundary position. Usually this is a good idea. There could be some case where the other isotope label data are not very constant, which is why there is an option to turn this off, but it defaults to On.
- I usually set Impute RT Outliers to On, if there is a smoothly varying trend of RT with acquisition time, or no RT drift, as this option could help the peak picking accuracy, by on the order of tenths of a percent. However there have been cases where experiments have some sharp change in retention time between replicates, where this option doesn't work as well as you might want it to.
- Press the Start button and the processing starts. On my laptop, it takes 1 minute to process the data for these 804 light/heavy precursors and 55 replicates.
- Note that we have not pressed the Send button yet, so Skyline still has its original integration boundaries.
- When the processing finishes, Expert Review ranks the peptides according to how many adjustments were made on the Experiment-level. This is a general measure of how big of a challenge the data posed. Right now, the first, or worst quality precursor is selected when the processing is finished.
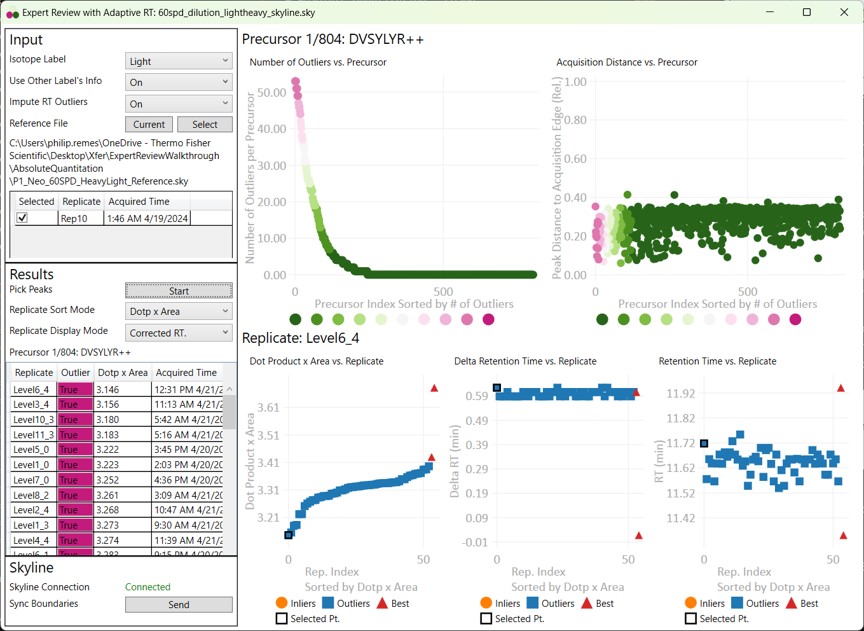
- Skyline is synced to Expert Review in a one-way mode, such that the active precursor in Expert Review is made active in Skyline, but not vice-versa.
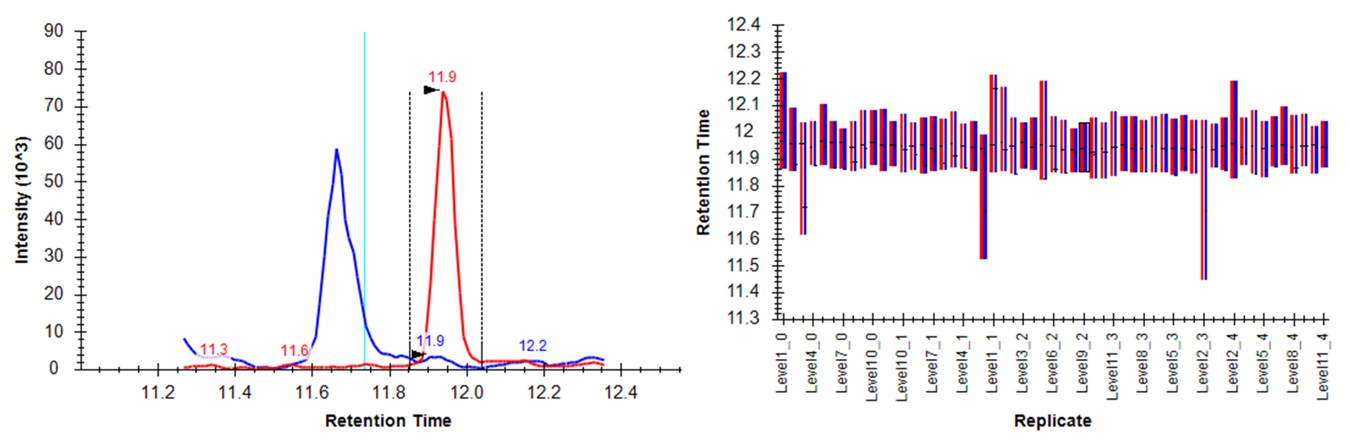
- Switch to the Skyline document to see the chromatograms for DVSYLYR, the one that has the most outliers. This is a challenging one for the light mode, because there is a huge endogenous peak at a retention time that is nearby to the heavy standard.
- Either press the Right key 3 times, or try to click the mouse on the 4th dot on the Number of Outliers vs Precursor graph. You can also zoom in on the plot with the mouse wheel, and double click to unzoom. You'll have highlighted the LTPTLYVGK precursor. This one has an endogenous time profile that is variable and does not present a clear pattern. It's no wonder that Skyline had a hard time here, using light boundaries.
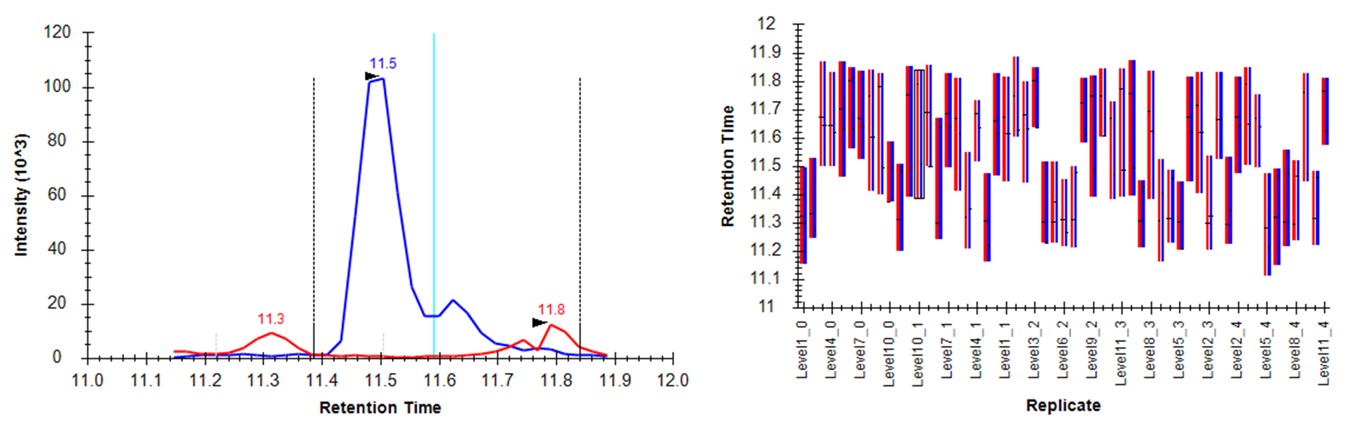
- Press the Send button. Skyline will take a few moments to import the Expert Review boundaries, and will stay on the LTPTLYVGK peptide. Now the boundaries are certainly more consistent, but they are in the wrong place, because the light data don't have a very good peak in the neighborhood of the actual heavy standard peak. The areas of the heavy peptides don't match with the expected dilution trend. This example shows again the perils of using light boundaries when there is no good light reference peak. If you go back to the DVSYLYR peptide, you'll see that here the results are not so satifactory either. In any event, Expert Review let us know which precursors were the most problematic. If you peruse the precursors that had fewer outliers corrected, the results are excellent. Let's see how things improve when we use the heavy peak boundaries.
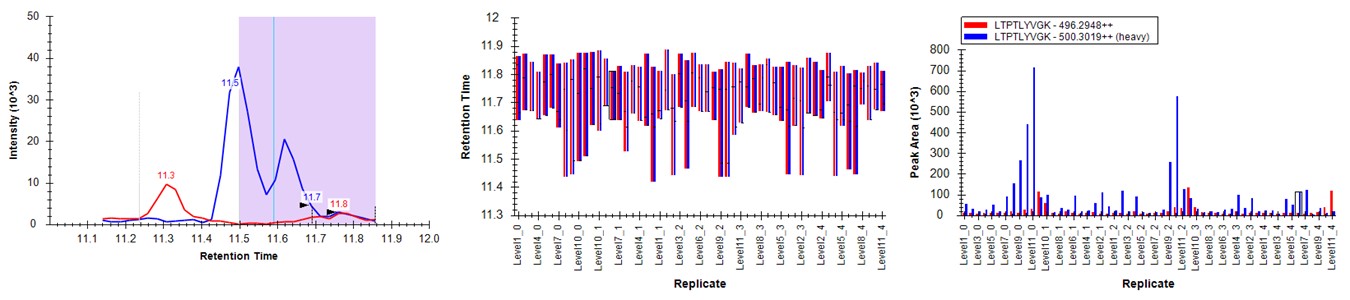
Expert Review - Heavy Boundary Mode
- Select Isotope Label Heavy, and Press the Start button. When the processing finishes, press the Send button. Notice that now the outliers plot shows that most precursors have many more replicates that needed Experiment-level correction. This makes sense, because every precursor is getting diluted below its limit of detection, eventually, and its replicate data no longer matches the reference.
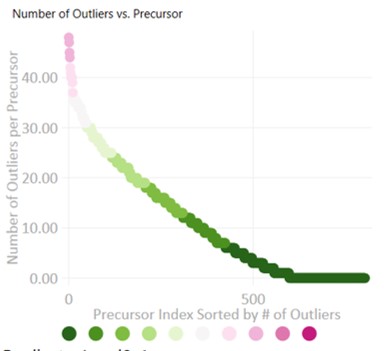
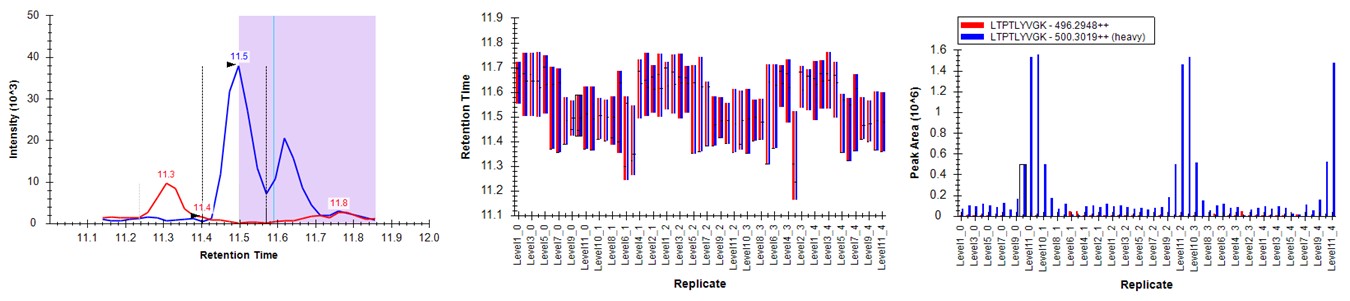
- Peruse the data a little bit, pressing the Right key on Expert review to look at the most challenging cases. They look good! In some cases, there are early eluting peaks with variable peaks that give poor results, or very nearby interferences that affect the absolute peak areas. There could be some improvements to be made in terms of making the integration boundary widths more consistent.
- If we look at the LTPTLYVGK example from before, the low level interference does move the boundary a little bit, but the areas of the peaks are much more in line with the dilution curve.
Conclusion
When performing dilutions of heavy standards in an endogenous background, use the heavy boundary mode, unless you are sure that all or most of the peptides have appreciable endogenous signal to use as references.
Label Free - E. Coli
Label Free - E. Coli
For this walkthrough, we will continue using data from our paper on Stellar MS with Adaptive RT and PRM Conductor. This section uses a dilution of E. Coli in a HeLa background.
- Open the file LabelFree / ecoli_small_dilution_skyline.sky.
- If you haven't installed the new PRM Conductor v1.1, do that
- Run Expert Review
- If you just did the absolute quantitation walkthrough, then Expert Review has the reference for that experiment, and is set to Isotope Label Heavy. We have to update this.
- Set Isotope Label to Not-Specified
- Press the Current Button. The reference grid is populated now with the replicates from this dilution. You could choose a replicate from this experiment to use as a reference, but I find it more convenient to use a separate reference.
- In the label free scenario, we have the advantage that we likely imported search results into Skyline, so all of the peaks likely are chosen correctly in some initial data set. The only problem is that the quality of these data may be low, due to the low injection times often used for a discovery DIA experiment.
- What I do is use the GPF as a reference to pick the peaks in PRM validation experiments, and then eventually use a PRM experiment as the reference for the remaining experiments in a study.
- Press the Select button, and choose LabelFree / ecolihela_gpf.sky. There are 6 LC injections that comprise this data set, but they were imported as a multi-injection replicate to Skyline, so they show up as a single replicate here. If you had imported the data as 6 single replicates, you could select all 6 of them in the reference grid here.
- Press Start to process the data, and then press the Send button to send the integration boundaries to Skyline.
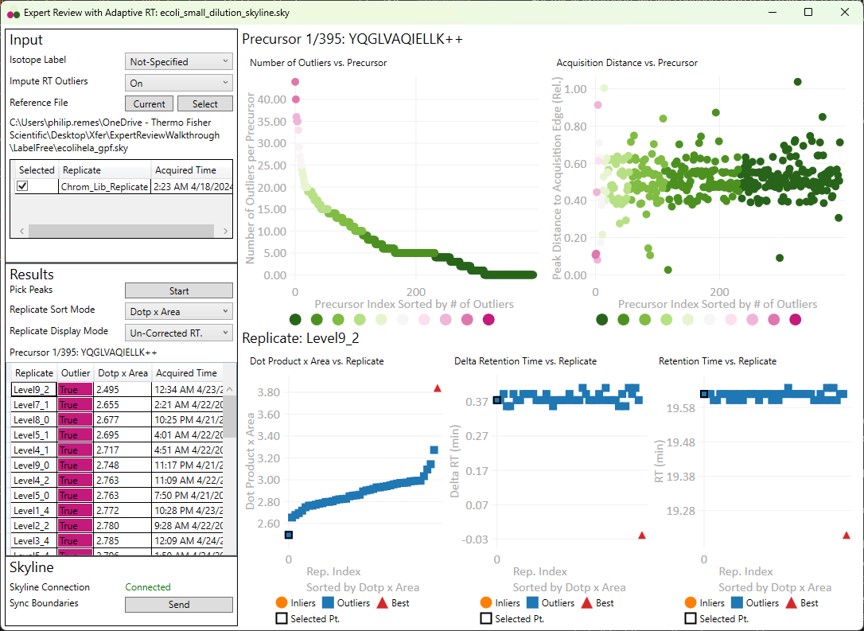
- Have a look at the worst precursors using the Right key. There are a few that should never have been included in the assay. Let's look at the worst precursor according to the number of outliers, YQGLVAQIELLK. Most of the integration boundaries are over near a small peak at the boundary. If we open the GPF Skyline document and find this peptide, we'll see that it has many more transitions.
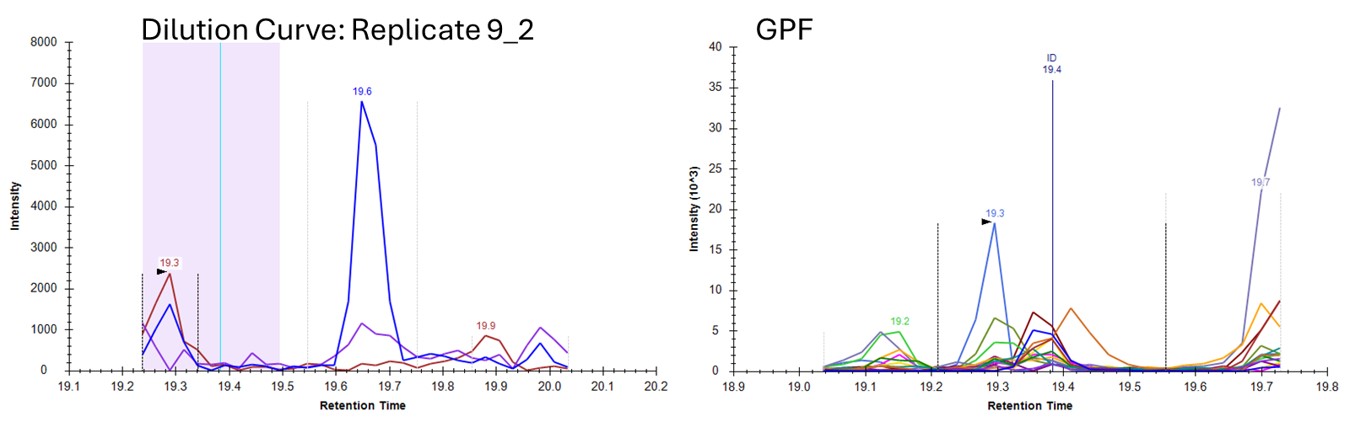
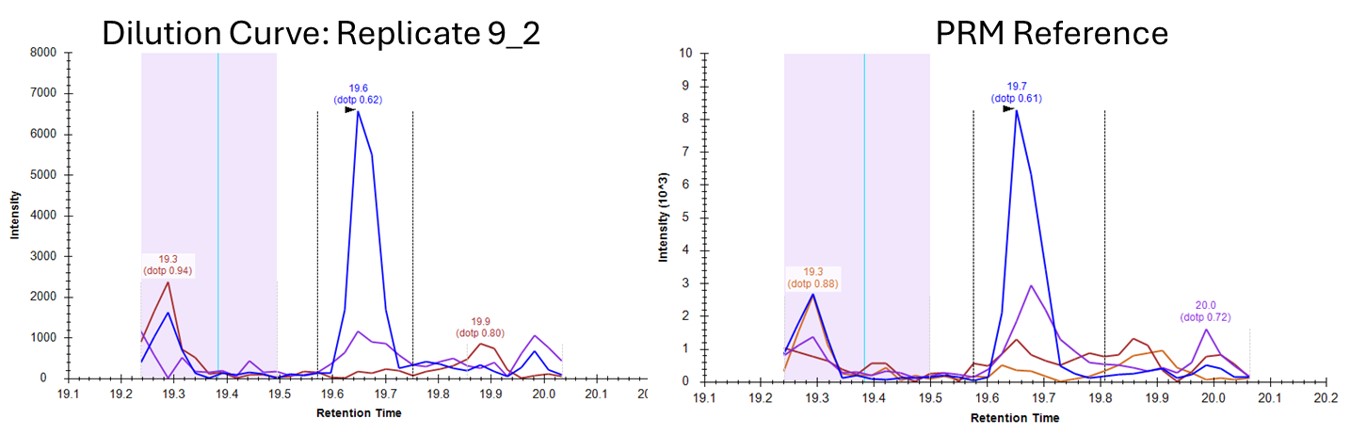
- Let's see if we can get better results by using a better reference, from PRM data. Press Select and choose the file ecoli_small_reference.sky, press Start, and then Send when the processing finishes. Use Ctrl+F and paste in YQGLVAQIELLK and search for it. Now these data look very nice, because the reference and dilution experiment data are very similar.
If you look in the Skyline Immediate Window log, you'll see some messages of the type:
2025-05-20 10:22:15.7649|WARN|Thermo.TNG.PeakPick.Algorithm.Logger|No reference found for precursor Sequence: QIIIATGEGAK
If you go to the reference file, for some reason this peptide is not found. It looks like there was an acquisition issue with this one, potentially caused by narrow acquisition windows, and an issue with Adaptive RT having a jump in its estimated retention time. We have since done some work to mitigate this kind of occurance in 1.1, so we hope that the low rate of such occurence will be even lower now.
- Now if we press the Right and Left keys in Expert Review to look through the precursors with the most outliers, they look good. There are a few precursors that we would consider dropping from the assay, or at least from the Skyline file, due to low S/N or bad peak shapes, but the peak integration boundaries look nice.
Conclusion
Expert Review can work with label free data too, but the similarity of the reference data to the later experiments is important. Another thing that we could have done to improve the results using the GPF reference, is to use PRM Conductor on it that file, and then make sure that the experiment data use the same transitions as the reference. That would have been what one normally would do, but these data were used to demonstrate the point. Since Stellar is a PRM machine, using the new transitions would just be a matter of reimporting the raw files in the Skyline documents.