Table of Contents |
guest 2025-09-17 |
Data Analysis Workflow
Sample Descriptions
| Replicate | Sample ID | Group | Acquisition Order | Collision Energy Profile |
| timsTOF_20230623_BSA-DSBU_S-Trap_pool_01_RD6_1_4677 | S-Trap Pool (quant) 1 | S-Trap Pool (quant) | 4677 | Low |
| timsTOF_20230623_BSA-DSBU_S-Trap_pool_02_RD6_1_4678 | S-Trap Pool (quant) 2 | S-Trap Pool (quant) | 4678 | Low |
| timsTOF_20230623_BSA-DSBU_S-Trap_pool_03_RD6_1_4679 | S-Trap Pool (quant) 3 | S-Trap Pool (quant) | 4679 | Low |
| timsTOF_20230623_BSA-DSBU_S-Trap_pool_04_RD6_1_4680 | S-Trap Pool (quant) 4 | S-Trap Pool (quant) | 4680 | Low |
| timsTOF_20230625_BSA-DSBU_S-Trap_pool_High_CE_01_RE1_1_4682 | S-Trap Pool (conditionaning) 1 | S-Trap Pool (conditionaning) | 4682 | High |
| timsTOF_20230625_BSA-DSBU_S-Trap_pool_Mid_CE_01_RE1_1_4683 | S-Trap Pool (conditionaning) 2 | S-Trap Pool (conditionaning) | 4683 | Mid |
| timsTOF_20230625_BSA-DSBU_S-Trap_pool_Low_CE_01_RE1_1_4684 | S-Trap Pool (conditionaning) 3 | S-Trap Pool (conditionaning) | 4684 | Low |
| timsTOF_20230625_BSA-DSBU_S-Trap_Rep_01_RE2_1_4686 | S-Trap Rep 1 | S-Trap Reps | 4686 | High |
| timsTOF_20230625_BSA-DSBU_S-Trap_Rep_02_RE3_1_4687 | S-Trap Rep 2 | S-Trap Reps | 4687 | High |
| timsTOF_20230625_BSA-DSBU_S-Trap_Rep_03_RE4_1_4688 | S-Trap Rep 3 | S-Trap Reps | 4688 | High |
| timsTOF_20230703_BSA-DSBU_S-Trap_NegCtrl_Rep_01_RD5_1_4722 | S-Trap NegCtrl Rep 1 | S-Trap NegCtrl Reps | 4722 | High |
| timsTOF_20230703_BSA-DSBU_S-Trap_NegCtrl_Rep_02_RD6_1_4723 | S-Trap NegCtrl Rep 2 | S-Trap NegCtrl Reps | 4723 | High |
| timsTOF_20230703_BSA-DSBU_S-Trap_NegCtrl_Rep_02_RD6_1_4723 | S-Trap NegCtrl Rep 3 | S-Trap NegCtrl Reps | 4724 | High |
| timsTOF_20230803_BSA-DSBU_SS_S-Trap_July2023Batch_DIA_HighCE_01_RD1_1_4837 | S-Trap DIA Rep 1 | S-Trap DIA | 4837 | High |
Short description of the MS files deposited in this Panorama repository.
Data Analysis Workflow
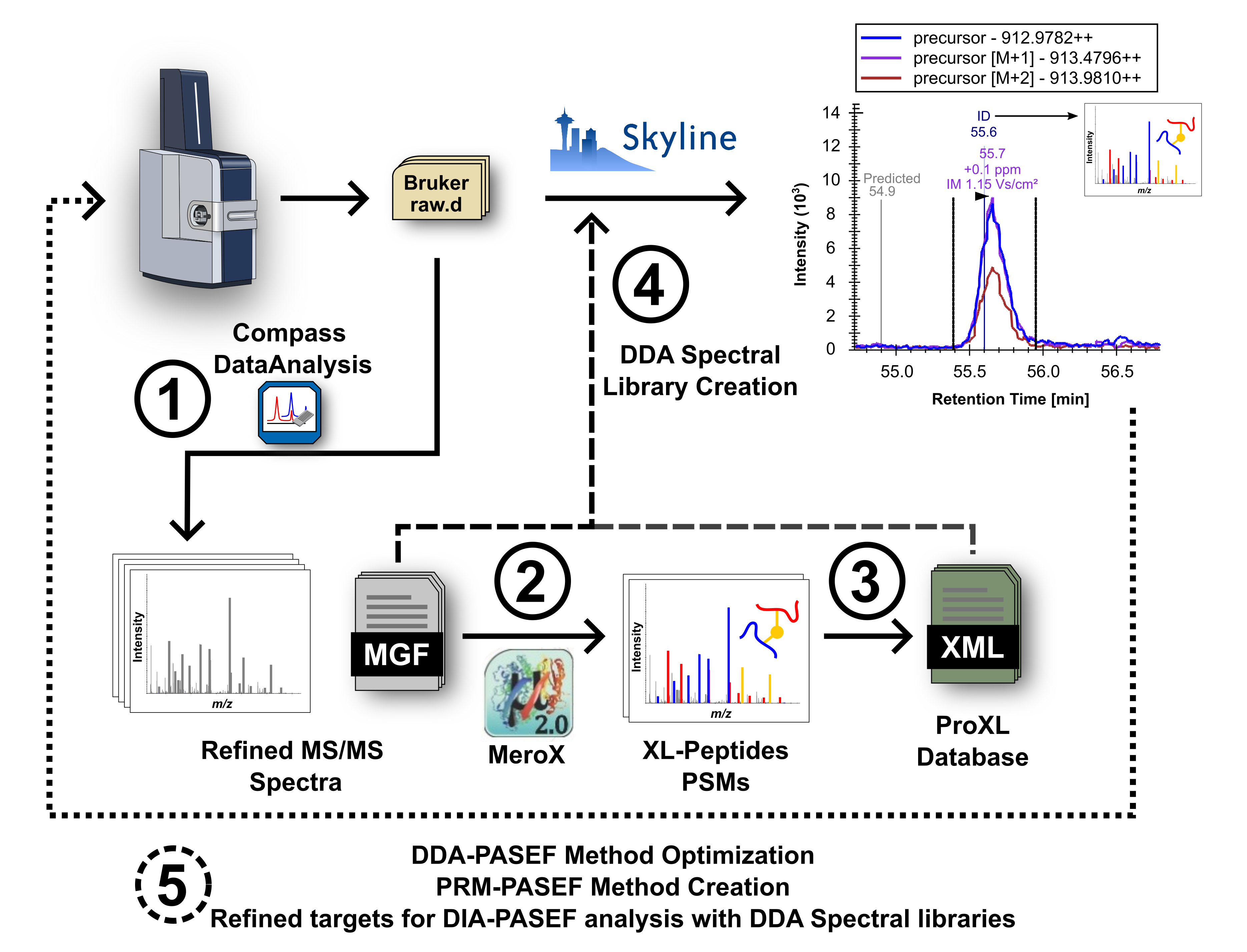
Data Analysis Workflow
1) Raw Bruker DDA-PASEF files are processed with DataAnalysis to obtain summed and filtered fragment ion spectra written in peak lists with a Mascot generic format. 2) The peak lists are then used for identification of cross-linked peptides with MeroX 2.0. 3) The native MeroX results are converted into a standard protein cross-linking (ProXL) XML database scheme. 4) The peak list files and the ProXL XML files are used to create a data dependent acquisition (DDA) spectral library. This spectral library is used to guide Skyline for the creation of extracted ion chromatograms (EICs) of the proposed cross-linked peptides. 5) The Skyline document is refined accounting for other analyte-specific properties (e.g.., isotopic pattern, peak shape, isotope co-elution, consistent chromatographic retention, etc.) accessible with Skyline. At this point data analysis can be stopped and report results or, alternatively, the refined documents can then be used to optimize DDA-PASEF experiments, design and export PRM-PASEF experiments, or serve as a template for analysis of DIA-PASEF data using DDA spectral libraries.