Factor H and related proteins in AMD
Cipriani V, Tierney A, Griffiths JR, Zuber V, Sergouniotis PI, Yates JRW, Moore AT, Bishop PN, Clark SJ, Unwin RD. Beyond factor H: The impact of genetic-risk variants for age-related macular degeneration on circulating factor-H-like 1 and factor-H-related protein concentrations. Am J Hum Genet. 2021 Aug 5;108(8):1385-1400. doi: 10.1016/j.ajhg.2021.05.015. Epub 2021 Jul 13. PMID: 34260948; PMCID: PMC8387294.
- Organism: Homo sapiens
- Instrument: 6495A Triple Quadrupole LC/MS
- SpikeIn:
Yes
- Keywords:
AMD, Factor H, FHL-1
-
Lab head: Richard Unwin
Submitter: Anna Tierney
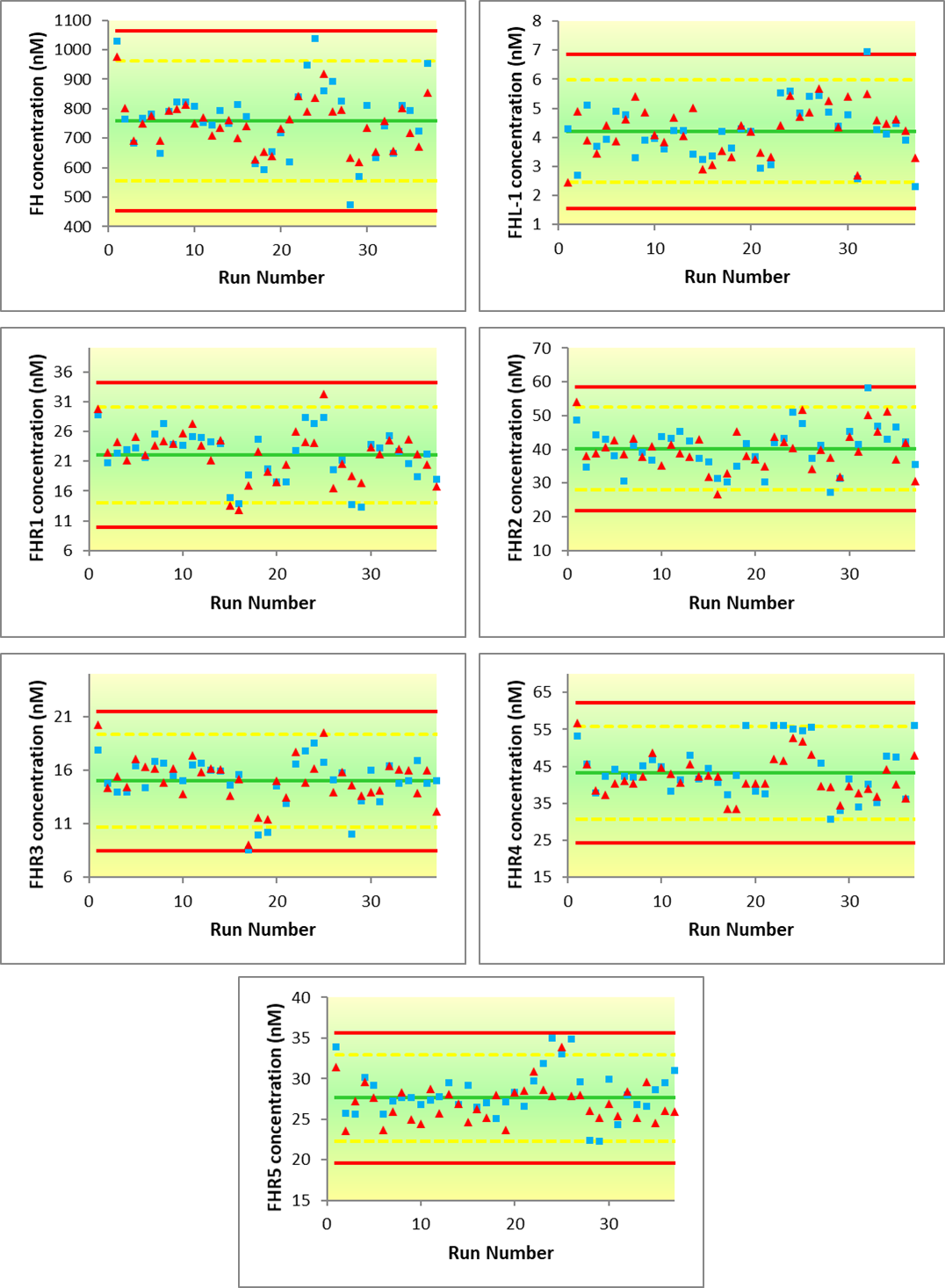
Age-related macular degeneration (AMD) is a common form of adult blindness. Risk of developing AMD is strongly linked to genetics, in particular at the Regulators of Complement Activation (RCA) locus on Chr1, which contains 6 genes, Complement Factor H (CFH) and Complement Factor H-related 1-5 (CFHR1-5). These encode seven distinct proteins, since CFH gives rise to both a full length Factor H (FH) and a small splice variant, FHL-1 protein. These proteins act as fluid-phase co-factors for the breakdown of C3b and hence regulation of the complement cascade. Many of these SNPs are intronic; there is speculation that they modify the systemic levels of these proteins, causing an imbalance on complement regulation which impacts AMD development. Since FH, Factor H-related (FHR1-5) and FHL-1 proteins exhibit a high degree of homology, there is no antibody-based assay available for the simultaneous quantification of all seven related proteins. Here we describe a novel mass spectrometry-based assay for the simultaneous detection and quantification of all seven highly homologous human complement proteins; FH, FHR1, FHR2, FHR3, FHR4, FHR5 and FHL-1, in plasma. Since proteolysis using trypsin is not amenable to the generation of reliable proteotypic peptides for all seven proteins, we utilised Endoproteinase Glu-C (V8 Protease) as an alternative. We were able to generate unique signature peptides for all seven of the target proteins in a simple protocol which requires no pre-fractionation or clean-up stages during sample preparation. The method described is shown to be capable of the robust, simultaneous detection and relative quantification of all seven proteins with linearity and limits of detection appropriate to physiological levels. Once validated, the method was successfully applied to a clinical set of AMD control samples and compared to an existing FH antibody-based approach.
SRM analyses of plasma digests were performed on a 6495 triple quadrupole mass spectrometer with iFunnel-equipped electrospray ion source (Agilent, Santa Clara, CA, USA) (Source parameters in Supplementary information Table 4) coupled to an Infinity 1200 Series liquid chromatography system consisting of 1290 autosampler, 1260 Quaternary Pump VL pump and TCC column oven modules (Agilent, Santa Clara, CA, USA). Samples were injected directly (4 μL, equivalent to approximately 28 μg peptides) onto a C18 column (250 mm x 2.1 mm I.D., Thermo Scientific Acclaim 120, 3 μm particle size) that was maintained at a temperature of 50 °C. Peptides were developed using a gradient elution of increasing acetonitrile concentration with Buffer A consisting of Water + 0.1 % formic acid and Buffer B being Acetonitrile + 0.1 % formic acid. The flow rate was maintained at 250 μL/min with an initial composition of 5 % Buffer B.
The following gradient elution profile was used to separate the peptides (time: %B): 0 min: 5 % B; 2 min: 5 % B; 3 min: 12 % B; 12 min: 15 % B; 15 min: 20 % B; 30 min: 25 % B; 31 min: 90 % B; 39 min:90 % B; 40 min: 5 % B; 49 min: 5 % B.
Plasma control samples were obtained from a case–control study (Cambridge AMD study) with subjects recruited from the southeast and northwest of England between 2002 and 2006. All affected subjects had CNV and/or GA. Controls were spouses, partners or friends of index patients. Blood samples were obtained at the time of interview. The EUGENDA created for clinical and molecular analysis of AMD comprises late AMD cases and controls recruited at Radboud University Medical Centre, the Netherlands, and University of Cologne, Germany. All participants provided written informed consent for clinical examination, epidemiological data collection and blood sampling for biochemical and genetic analyses. Serum and plasma samples were used for all measurements. Donor eye tissue was obtained from Manchester Eye Tissue Repository (ethically approved Research Tissue Bank, UK NHS Health Research Authority ref. 15/NW/0932). The banked tissue was collected and stored within 48h of death; there was prior informed consent for research use. Human Tissue Act 2004 (UK) guidelines were followed. For all studies, ethical approval was obtained from either national or local ethics committees (NRES Committee East Midlands – Derby for the Cambridge AMD study; Arnhem–Nijmegen Commissie Mensgebonden Onderzoek (CMO) and Ethics Commission of Cologne University’s Faculty of Medicine for EUGENDA) and adhered to the tenets of the Declaration of Helsinki.
Frozen plasma samples were allowed to thaw to room temperature before being vortexed hard for 5 min to dissolve any soluble material, then centrifuged at 13,300g for 30 min to settle any insoluble material. A 5 μL aliquot was carefully transferred to a 1.5 mL LoBind Eppendorf tube for processing. Care was taken to avoid transferring any lipid-like material which may have risen to the surface.
To the 5 μL plasma aliquot, 90 μL of 50 mM ammonium bicarbonate (pH 7.8), 2 μL of ProteaseMAX™ (Promega, Southampton, UK) solution (1% w/v in 50 mM ammonium bicarbonate) and 1 μL of 500 mM dithiothreitol prepared in 50 mM ammonium bicarbonate was added. This was then vortexed briefly to mix, then given a pulse spin before incubating at 56 °C for 25 min.
After cooling to room temperature, 3 μL 500 mM iodoacetamide (prepared in 50 mM ammonium bicarbonate) was added. This was vortexed briefly to mix, then given a pulse spin before incubating at room temperature and in the dark for 15 min.
To digest the protein, a further 43 μL of 50 mM ammonium bicarbonate (pH 7.8), 1 μL of ProteaseMAX solution (1% w/v in 50 mM ammonium bicarbonate) and 5 μL of 1 μg/μL endoproteinase Glu-C (Roche, Mannheim, Germany) were added, the tube was vortexed briefly to mix, then given a pulse spin before incubating for 16 hours at 25 °C with shaking (400 rpm).
Spiking solution was prepared immediately prior to sample addition by adding 195 μL 50:50 acetonitrile:water to a 5 μL aliquot of the mixed SIS solution. 2 μL of this was carefully added to each digested sample along with 6 μL 10 % v/v TFA, vortexed briefly to mix, then pulse spin. It was next placed into a centrifugal evaporator (Eppendorf) at 45 °C and taken to dryness. Finally, the dried peptides were reconstituted in 50 μL 0.1 % TFA and vortexed to dissolve any residue before centrifuging at 13,300g for 30 min to settle any insoluble/particulate material. Approximately 48 μL (taking care to leave behind any precipitated material) was transferred to a LC autosampler vial for subsequent analysis by LC-MS/MS.
Created on 1/7/21, 5:41 PM