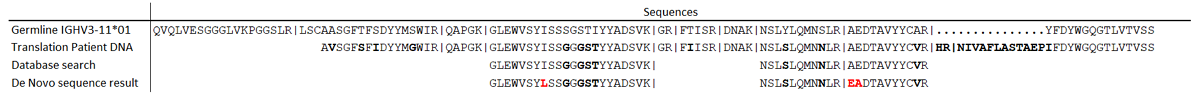Tables |
2024-05-29 |
Additional data files has been uploaded to the RAW data section of the repository.
Patient and sample characteristics
More detailed clinical information is available in a separate document
| Patient number | M-protein | SPEP fraction | Days follow-up a | Samples measured by MSb |
| 1 | IgA lambda | beta | 1422 | 33 |
| 2 | IgG kappa | gamma | 584 | 12 |
| 3 | IgG kappa | gamma | 760 | 31 |
| 4 | IgA lambda | gamma | 1512 | 30 |
| 5 | IgG kappa | gamma | 1707 | 36 |
| 6 | IgA kappa | gamma | 702 | 26 |
| 7 | IgG kappa | gamma | 918 | 17 |
| 8 | IgG kappa | gamma | 1799 | 32 |
| 9 | IgG kappa | gamma | 1914 | 28 |
anumber of days between the first and the last time point in the disease, measured by mass spectrometry (MS)
beach sample represents serum of a distinct time point in the disease course, analyzed by serum protein electrophoresis (SPEP) and excised for MS measurement,
De novo sequence results for the selected multiple myeloma patients.
| Patient number | M-protein chain | De novo M-protein peptide sequence | ALC % c | Germline peptide sequence |
| 1 | heavy | DSVFLQMNSLR b | 73 | NSLYLQMNSLR |
| 2 | heavy | DGQLVESGGGSAQPGGSLR a | 86 | EDQLVESGGGLVQPGGSLR |
| 3 | heavy | LSCEASGFTFR | 91 | LSCAASGFTF |
| 4 | heavy | SPPVSVSHVEANSPGQTASLTCSGDK | 72 | SYELTQLPSVSVSPGQTARITCSGDV |
| 5 | heavy | MTNMDPVDTATYYCARb | 76 | MTNMDPVDTATYYCAR d |
| 5 | light | LVLTQSPATLSLSASPNAAK a | 73 | IVLTQSPATLSLSP |
| 6 | heavy | VELLVESGGDLVQPGGSLR | 82 | LVESGGGLVQPGGSLR |
| 6 | light | LEVLTQSPGTLSLSPDAR | 82 | VLTQSPGTLSLSPGER |
| 7 | heavy | EDTALFYCVK | 84 | EDTAVYYCVK |
| 7 | light | DLQMTQSPSSLSASVGDK b | 78 | DIQMTQSPSSLSASVGDR |
| 8 | heavy | SMTAADTGVYYCAR | 80 | SVTAVDTGVYYCAR |
| 8 | light | QLDLTQSPSSLSASVGDR | 72 | IQLTQSPSSLSASVGDR |
| 9 | light | LSLYGASNLQGGVPSK a | 76 | LLIYSASNLQSGVPSR |
a de novo sequences derived from targeted measurements is shown, with differences to DDA based data
b The peptide was monitored with all methionine residues in the oxidized form
c ALC = average local confidence score from PEAKS
d The germline peptide lacks an N-terminal tryptic site.
Sequence, m/z, charge states and fragment types of selected patient-specific M-protein peptides used for M-protein monitoring with targeted mass spectrometry.
| Patient | Sequence | m/z | z | Fragments |
| 1 | DSVFLQM[Ox]NSLR | 663.3295 | 2 | y5-8 |
| 2 | DGQLVESGGGSAQPGGSLR | 886.4321 | 2 | y6; y11-15 |
| 3 | LSC[CAM]EASGFTFR | 637.7953 | 2 | y5-10 |
| 4 | SPPVSVSHVEANSPGQTASLTC[CAM]SGDK | 871.4134 | 3 | y6; y8-10; y13 |
| 5 | M[Ox]TNM[Ox]DPVDTATYYC[CAM]AR | 970.8948 | 2 | y4; y6-9; y11 |
| 6 | VELLVESGGDLVQPGGSLR | 963.0206 | 2 | y6-9; y11-14 |
| 7 | EDTALFYC[CAM]VK | 623.2946 | 2 | y4-8 |
| 8 | SM[Ox]TAADTGVYYC[CAM]AR | 791.3372 | 2 | y5; y7-12 |
| 9 | LSLYGASNLQGGVPSK | 795.9279 | 2 | y3, y6, y10-13 |
[Ox] = oxidation
[CAM] = carbamidomethylation
DNA and de novo sequencing results of the reference patient.
Differences between the patient specific M-protein and the most homologous germline sequence are bold. Differences between the de novo sequencing result and the DNA-derived sequence are in red. No light chain DNA data was available to validate sequences obtained from de novo sequencing for the light chain. Trypsin cleavage sites are indicated with | symbols.